HDV
Rfam ID: RF00094
click into different sections:
Timeline
-
1988 Discovery[1]
-
1988 Discovery[2]
-
1989 Discovery[3]
-
1990 84 nucleotides are required for rapid and efficient self-cleavage[4]
-
1991 Pseudoknot-like secondary structure[6]
-
1992 The P4 duplex can reduce the minimum size to about 65 nucleotides[7]
-
1993 Nonspecifific divalent cations are required for self-cleavage[8]
-
1996 Use of cis-delta ribozyme generated 3'homogeneous RNA ends[9]
-
1997 NMR structure of the isolated central hairpin(Stem Loop Ⅲ)[10]
-
1998 Crystal structure[11]
-
2000 C75 acts as the general acid and ribozyme-bound hydrated metal hydroxide as the general base [12]
-
2004 Precursor structures [13]
-
2005 It is proved that C75 acts as the general acid[14]
-
2010 Precleavage structures[15]
-
2015 The HDV ribozyme variants were discovered [16]
-
2015 Transition state features [17]
-
2016 Dynamic reaction mechanism model with two Mg2+ ions [18]
-
2019 Double-pseudoknot HDV can self-cleavge with the same mechanism as the WT ribozyme[20]
Description
The hepatitis delta virus (HDV) ribozyme is a non-coding RNA found in the hepatitis delta virus that is necessary for viral replication. It is the only known human virus that utilizes ribozyme activity to infect its host. The ribozyme acts to process the RNA transcripts to unit lengths in a self-cleavage reaction during replication of the hepatitis delta virus, which is thought to propagate by a double rolling circle mechanism. The ribozyme is active in vivo without any protein factors and was the fastest known naturally occurring self-cleaving RNA at the time of its discovery.
Structure and mechanism
2D representation
Secondary structure depictions of the HDV ribozyme with the scissile phosphates (blue arrow )and the general acid (red circle ) and base (blue dot) are shown. The HDV ribozymes act with the direct involvement of metal ions shown by green spheres with octahedral coordination of red water molecules of hydration.

|
3D visualisation
The overall structure of the HDV ribozyme was generated from PDB ID:1DRZ at 2.3 Å resolution.
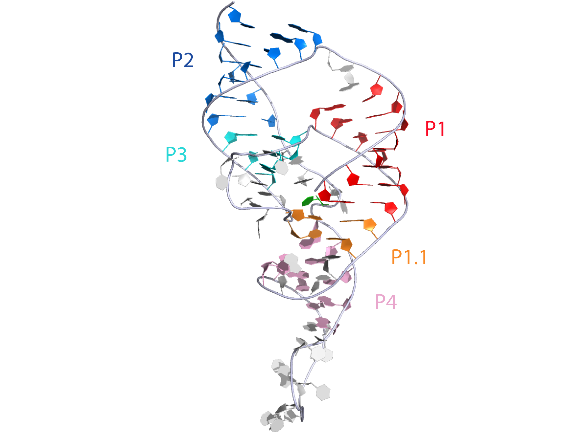 |
|
Catalytic centre
Close view of the catalytic centre of the HDV ribozyme, showing the proximity of C75. This image was drawn using PDB ID: 4PRF at 2.40 Å resolution.(left) Proposed mechanisms for the HDV ribozyme. The HDV ribozyme uses general acid-base catalysis, in which the general base is a metal ion-bound hydroxide ion, and cytosine N3 as the general acid.(right)
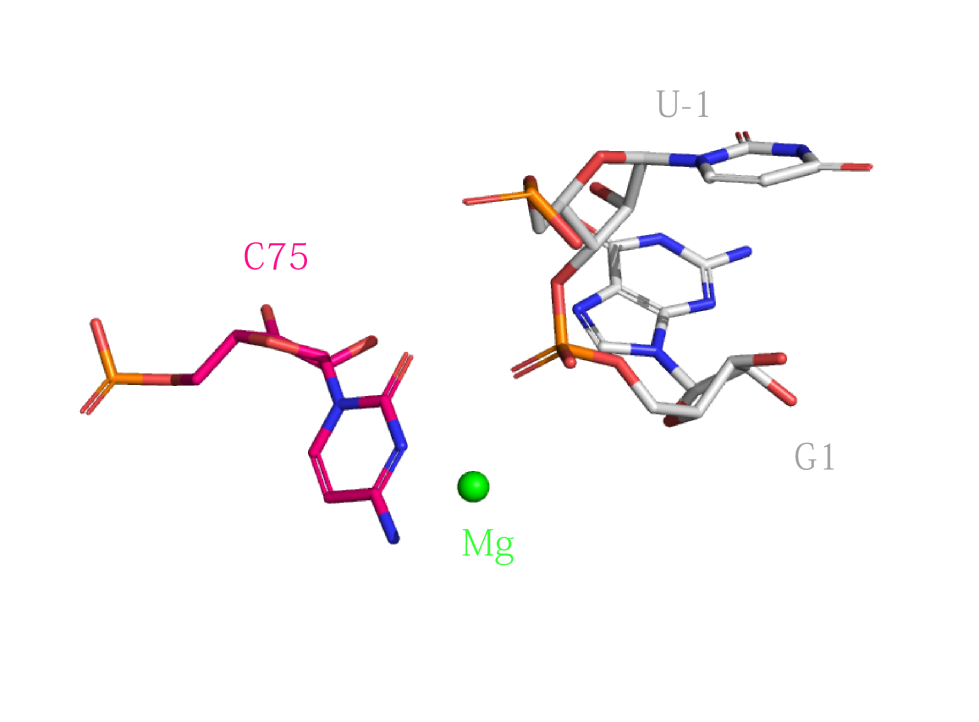 |
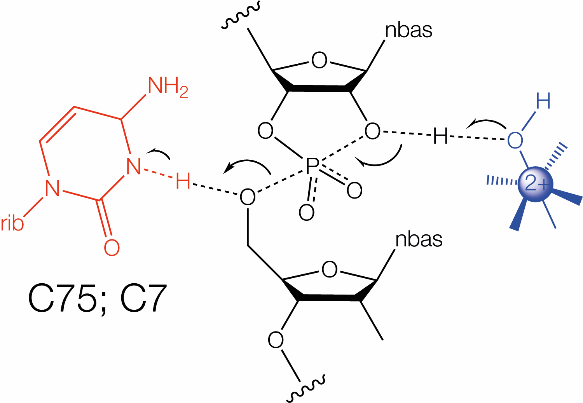 |
Chemical mechanism
Unlike many of the nucleolytic ribozymes, the HDV ribozyme is essentially inactive in both 4M Li+ and Co(III) hexammine ions[12] suggesting a direct role for the divalent metal ions. Yet the rate of cleavage by the HDV ribozyme is pH dependent, indicating proton transfer in the transition state, consistent with general acid–base catalysis. The rate of cleavage by the genomic HDV ribozyme was found to increase in a log-linear manner with pH with gradient of 1, up to pH = 6.5. Thereafter the rate remained at a plateau with further increase in pH. The data fitted a pKa = 6.1 that might correspond to a cytosine in an electronegative environment. The other participant would then have to have a high pKa and this might correspond to a hydrated metal ion. The plateau would then correspond to complete deprotonation of cytosine if it were acting as a general base. Alternatively, the plateau might be due to compensatory simultaneous deprotonation of cytosine acting as the general acid and a metal hydroxide acting as the general base. Bevilacqua and coworkers [12]repeated the pH titration of reaction rate in the absence of divalent cations, with 1M Na+ ions to ensure ribozyme folding, whereupon the pH dependence inverted. The cleavage rate was maximal at low pH and declined linearly with increase in pH, corresponding to a pKa = 5.7. Under these conditions, water takes the role of the hydrated metal ion, isolating the titration of C75. Since the cleavage rate reduced with increasing pH, this indicated that C75 acts as the general acid. A definitive assignment of the general acid was made by Das and Piccirilli [14]. 5'-phosphorothiolate substitution resulted in almost full restoration of activity of the antigenomic HDV C76U ribozyme. Furthermore, the pH dependence of this modified ribozyme then only reflected the deprotonation of the base, because the deprotonation of the general acid was no longer kinetically significant. In addition, C76 N3C (3-deazacytosine) had undetectable levels of activity, but cleavage was restored by 5'-phosphorothiolate substitution. Collectively the data suggest that HDV uses general acid-base catalysis mediated by a metal hydroxide as the general base and cytosine N3 as the general acid in the cleavage reaction.Proposed mechanisms for twister ribozyme using guanine and adenine nucleobases as general base and acid, respectively, in their cleavage reactions (A7 corresponds to A1, G33 corresponds to G45)
References
[1] Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus.
Kuo, M. Y., L. Sharmeen, G. Dinter-Gottlieb and J. Taylor
J Virol 62(12): 4439-4444.(1988)
[2] Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage.
Sharmeen, L., M. Y. Kuo, G. Dinter-Gottlieb and J. Taylor
J Virol 62(8): 2674-2679.(1988)
[3] Human hepatitis delta virus RNA subfragments contain an autocleavage activity.
Wu, H. N., Y. J. Lin, F. P. Lin, S. Makino, M. F. Chang and M. M. Lai
Proceedings of the National Academy of Sciences 86(6): 1831-1835.(1989)
[4] The self-cleaving domain from the genomic RNA of hepatitis delta virus: sequence requirements and the effects of denaturant.
Perrotta, A. T. and M. D. Been
Nucleic Acids Res 18(23): 6821-6827.(1990)
[5] Evidence that genomic and antigenomic RNA self-cleaving elements from hepatitis delta virus have similar secondary structures.
Rosenstein, S. P. and M. D. Been
Nucleic Acids Research 19(19): 5409-5416.(1991)
[6] A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA.
Perrotta, A. T. and M. D. Been
Nature 350(6317): 434-436.(1991)
[7] Secondary structure of the self-cleaving RNA of hepatitis delta virus: applications to catalytic RNA design.
Been, M. D., A. T. Perrotta and S. P. Rosenstein
Biochemistry 31(47): 11843-11852.(1992)
[8] Self-cleavage activity of the genomic HDV ribozyme in the presence of various divalent metal ions.
Suh, Y. A., P. K. Kumar, K. Taira and S. Nishikawa
Nucleic Acids Res 21(14): 3277-3280.(1993)
[9] Use of cis- and trans-ribozymes to remove 5' and 3' heterogeneities from milligrams of in vitro transcribed RNA.
Ferre-D’Amare, A. R. and J. A. Doudna
Nucleic Acids Res 24(5): 977-978(1996)
[10] The structure of the isolated, central hairpin of the HDV antigenomic ribozyme: novel structural features and similarity of the loop in the ribozyme and free in solution.
M H Kolk , H A Heus and C W Hilbers
The EMBO Journal 16(12): 3685-3692.(1997)
[11] Crystal structure of a hepatitis delta virus ribozyme.
Ferré-D’Amaré, A. R., K. Zhou and J. A. Doudna
Nature 395(6702): 567-574.(1998)
[12] General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme.
Nakano, S., D. M. Chadalavada and P. C. Bevilacqua
Science 287(5457): 1493-1497.(2000)
[13] A conformational switch controls hepatitis delta virus ribozyme catalysis.
Ke, A., K. Zhou, F. Ding, J. H. Cate and J. A. Doudna
Nature 429(6988): 201-205.(2004)
[14] General acid catalysis by the hepatitis delta virus ribozyme.
Das, S. R. and J. A. Piccirilli
Nat Chem Biol 1(1): 45-52.(2005)
[15] A 1.9 A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage.
Chen, J. H., R. Yajima, D. M. Chadalavada, E. Chase, P. C. Bevilacqua and B. L. Golden
Biochemistry 49(31): 6508-6518.(2010)
[16] New classes of self-cleaving ribozymes revealed by comparative genomics analysis.
Weinberg, Z., P. B. Kim, T. H. Chen, S. Li, K. A. Harris, C. E. Lünse and R. R. Breaker
Nature Chemical Biology 11(8): 606-610.(2015)
[17] Transition State Features in the Hepatitis Delta Virus Ribozyme Reaction Revealed by Atomic Perturbations.
Koo, S. C., J. Lu, N. S. Li, E. Leung, S. R. Das, M. E. Harris and J. A. Piccirilli
J Am Chem Soc 137(28): 8973-8982.(2015)
[18] A Two-Metal-Ion-Mediated Conformational Switching Pathway for HDV Ribozyme Activation.
Lee, T. S., B. K. Radak, M. E. Harris and D. M. York
ACS Catal 6(3): 1853-1869.(2016)
[19] Classification of the nucleolytic ribozymes based upon catalytic mechanism.
Lilley, D. M. J.
F1000Res 8.(2019)
[20] Design of highly active double-pseudoknotted ribozymes: a combined computational and experimental study.
Yamagami, R., M. Kayedkhordeh, D. H. Mathews and P. C. Bevilacqua
Nucleic Acids Res 47(1): 29-42.(2019)
 Home
Home Database
Database Research
Research About us
About us

