VS ribozyme
Rfam ID:na
click into different sections:
Timeline
-
1990 Discovery[1]
-
1995 Secondary structure[2]
-
2001 Importance of the 2-3-6 helical junction[3]
-
2001 A730 loop is important[4]
-
2002 A756 is critical for catalysis[7]
-
2007 G638 is critical for catalysis[10]
-
2008 SAXS-deriverd structure[11]
-
2009 The soixante-neuf experiment[12]
-
2010 Catalytic mechanism[13]
-
2011null Detailed discussion of the chemical mechanism[14]
-
2015 Crystal structure[18]
-
2020 Additional experiments to summarize the structure and function of VS ribozyme[20]
Description
Varkud satellite (VS) ribozyme is the largest known nucleolytic ribozyme and found to be embedded in VS RNA. VS RNA is a long non-coding RNA exists as a satellite RNA and is found in mitochondria of Varkud-1C and few other strains of Neurospora. VS ribozyme contains features of both catalytic RNAs and group 1 introns. VS ribozyme has both cleavage and ligation activity and can perform both cleavage and ligation reactions efficiently in the absence of proteins. VS ribozyme undergo horizontal gene transfer with other Neurospora strains. VS ribozymes have nothing in common with other nucleolytic ribozymes.
Structure and mechanism
2D representation
Secondary structure of the VS ribozyme, the general acid A756 and general base G638 are shown in red and blue respectively.

|
3D visualisation
Crystal structure of the VS ribozyme, his representation was generated from PDB ID: 4R4V at 3.07Å resolution. The ribozyme crystallized was a G638A mutant used to prevent self-cleavage occurring during crystallization.
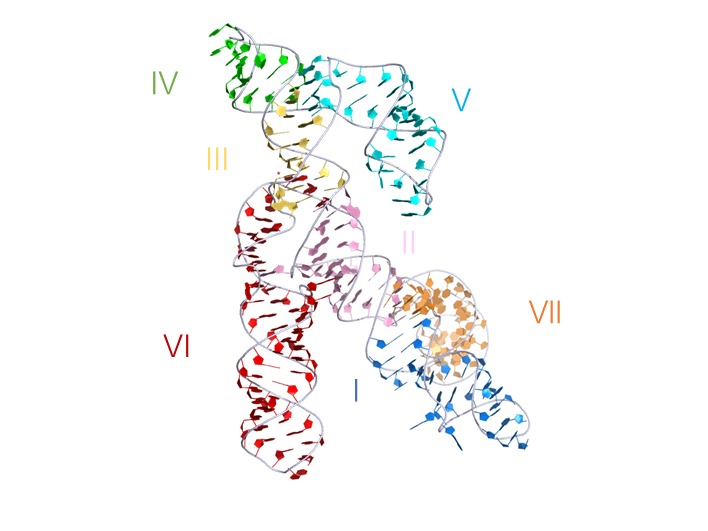 |
|
Catalytic centre
The active center of the ribozyme, he key catalytic participants are the nucleobases of G638 (replaced by adenine in this structure) and A756, the general base and acid, respectively, in the cleavage reaction (PDB ID: 4R4V).
Schematic of biochemically inferred mechanism of catalysis by the VS ribozyme involving general acid-base catalysis by G638 and A756. Arrows indicate bonds that are formed or broken during the transition state.
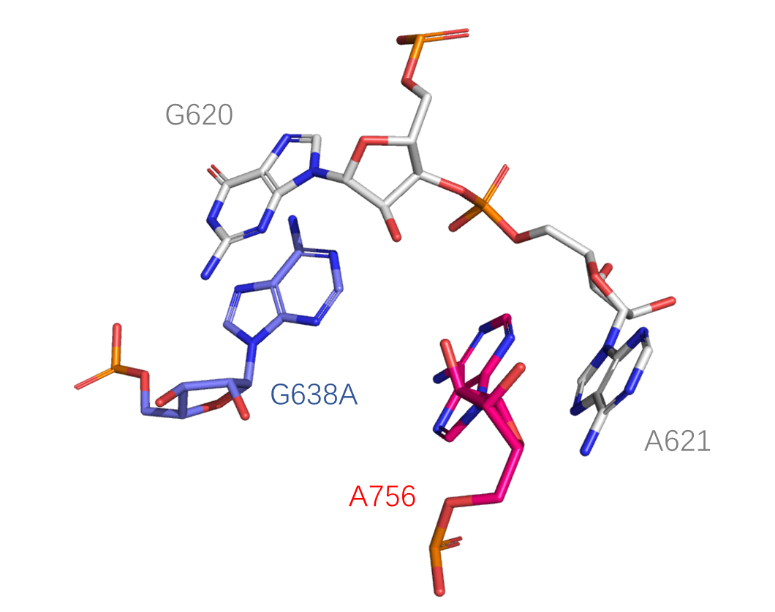 |
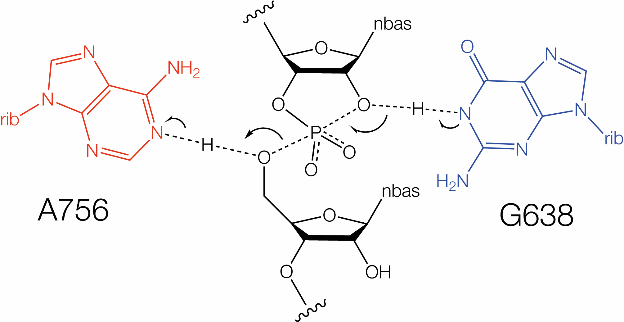 |
Chemical mechanism
The VS ribozyme employs general acid-base catalysis, using the nucleobases of G638 (in its deprotonated form)[10] as general base to remove the proton from the 2’-OH nucleophile and A756 (in its protonated form)[4,7] as the general acid to protonate the leaving group. The pH - rate profile for the VS ribozyme is bell shaped, fitting pKa values of 5.2 and 8.4 corresponding to A756 and G638 respectively [10]. The acid and base were assigned by 5’-phosphorothiolate substitution experiments [13]. The pH dependence of a VS G638DAP ribozyme cleavage reaction indicated that proton transfer contributes at least 102-103 fold to the catalytic power of the ribozyme[10]. When the crystal structure of the VS ribozyme was eventually solved it was found that G638 and A756 were adjacent to the O2’ and O5’ atoms respectively, consistent with their proposed roles in catalysis[18]. Note that despite different overall RNA folds, the catalytic mechanisms of the VS and hairpin ribozymes are closely similar, both using G (general base) + A (general acid) mechanisms.
References
[1] A site-specific self-cleavage reaction performed by a novel RNA in neurospora mitochondria.
Saville, B. J. and R. A. Collins
Cell 61(4): 685-696.(1990)
[2] A secondary-structure model for the self-cleaving region of Neurospora VS RNA.
Beattie, T. L., J. E. Olive and R. A. Collins
Proc Natl Acad Sci U S A 92(10): 4686-4690.(1995)
[3] Structure, folding and activity of the VS ribozyme : Importance of the 2-3-6 helical junction
D.A. Lafontaine, D.G. Norman and D.M.J. Lilley
EMBO J. 20 1415-1424 (2001)
[4] The A730 loop is an important component of the active site of the VS ribozyme.
Lafontaine, D. A., T. J. Wilson, D. G. Norman and D. M. Lilley
J Mol Biol 312(4): 663-674.(2001)
[5] A pH controlled conformational switch in the cleavage site of the VS ribozyme substrate RNA.
Flinders, J. and T. Dieckmann
J Mol Biol 308(4): 665-679.(2001)
[6] The global structure of the VS ribozyme.
D.A. Lafontaine, D.G. Norman and D. M.J. Lilley
EMBO J. 21, 2461-2471(2002)
[7] Functional Group Requirements in the Probable Active Site of the VS Ribozyme.
Lafontaine, D. A., T. J. Wilson, Z.-Y. Zhao and D. M. J. Lilley
Journal of Molecular Biology 323(1): 23-34.(2002)
[8] Efficient, pH-dependent RNA ligation by the VS ribozyme in trans
A.C. McLeod and D.M.J. Lilley
Biochemistry 43, 1118 – 1125(2004)
[9] Nuclear magnetic resonance structure of the Varkud satellite ribozyme stem-loop V RNA and magnesium-ion binding from chemical-shift mapping.
Campbell, D. O. and P. Legault
Biochemistry 44(11): 4157-4170.(2005)
[10] A guanine nucleobase important for catalysis by the VS ribozyme.
Wilson, T. J., A. C. McLeod and D. M. Lilley
EMBO J 26(10): 2489-2500.(2007)
[11] The complete VS ribozyme in solution studied by small-angle X-ray scattering.
Lipfert, J., J. Ouellet, D. G. Norman, S. Doniach and D. M. Lilley
Structure 16(9): 1357-1367.(2008)
[12] Formation of an active site in trans by interaction of two complete Varkud Satellite ribozymes
J. Ouellet, M. Byrne and D. M. J. Lilley
RNA 15, 1822-1826(2009)
[13] Nucleobase-mediated general acid-base catalysis in the Varkud satellite ribozyme.
Wilson, T. J., N. S. Li, J. Lu, J. K. Frederiksen, J. A. Piccirilli and D. M. Lilley
Proc Natl Acad Sci U S A 107(26): 11751-11756.(2010)
[14] Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid-base catalysis ? A critical assessment of available experimental data.
T. J. Wilson and D. M. J. Lilley.
RNA 17, 213-221 (2011)
[15] NMR structure of the A730 loop of the Neurospora VS ribozyme: insights into the formation of the active site.
Desjardins, G., E. Bonneau, N. Girard, J. Boisbouvier and P. Legault
Nucleic Acids Res 39(10): 4427-4437.(2011)
[16] Nuclear magnetic resonance structure of the III-IV-V three-way junction from the Varkud satellite ribozyme and identification of magnesium-binding sites using paramagnetic relaxation enhancement.
Bonneau, E. and P. Legault
Biochemistry 53(39): 6264-6275.(2014)
[17] The NMR structure of the II-III-VI three-way junction from the Neurospora VS ribozyme reveals a critical tertiary interaction and provides new insights into the global ribozyme structure.
Bonneau, E., N. Girard, S. Lemieux and P. Legault
RNA 21(9): 1621-1632.(2015)
[18] Crystal structure of the Varkud satellite ribozyme.
Suslov, N. B., S. DasGupta, H. Huang, J. R. Fuller, D. M. Lilley, P. A. Rice and J. A. Piccirilli
Nat Chem Biol 11(11): 840-846.(2015)
[19] Structural Basis for Substrate Helix Remodeling and Cleavage Loop Activation in the Varkud Satellite Ribozyme.
DasGupta, S., N. B. Suslov and J. A. Piccirilli
J Am Chem Soc 139(28): 9591-9597.(2017)
[20] Confluence of theory and experiment reveals the catalytic mechanism of the Varkud satellite ribozyme.
Ganguly, A., B. P. Weissman, T. J. Giese, N. S. Li, S. Hoshika, S. Rao, S. A. Benner, J. A. Piccirilli and D. M. York
Nat Chem 12(2): 193-201.(2020)
 Home
Home Database
Database Research
Research About us
About us

