Hairpin
Rfam ID: RF00173
RF04190
RF04191
click into different sections:
Timeline
-
1986 Discovery[1]
-
1993 Sequence/Secondary structure [2]
-
2001 Essential role of an active-site G8 in hairpin ribozyme catalysis[6]
-
2002 Crystal structure[7]
-
2005 Essential role of an active-site A38 in hairpin ribozyme catalysis[8]
-
2006 Crystal structure[9]
-
2012 Catalytic mechanism[10]
-
2019 Engineering of hairpin ribozyme variants[11]
-
2021 Situ activation of RNA substrates under reaction conditions amenable to catalysis by the hairpin ribozyme[12]
-
2021 Expand the number of natural hairpin ribozymes[13]
-
2022 ViroidDB: a database of viroids and viroid-like circular RNAs[14]
-
2022 Engineering of hairpin ribozyme variants[15]
Description
The GlmS ribozyme is predominantly present in gram-positive bacteria, and it is a riboswitch that inhibits the synthesis of glucosamine-6-phosphate. In addition, GlmS riboswitch is a self-cleft ribozyme located in the 5' untranslated region of the GlmS gene. Study on the structure and function of GlmS riboswitch will be beneficial to develop new targets for antibiotic action.
Structure and mechanism
2D representation
Secondary structure depictions of the nucleolytic ribozymes with the scissile phosphates (cyan) arrowed, and the general acid (red) and base (blue) shown.

|
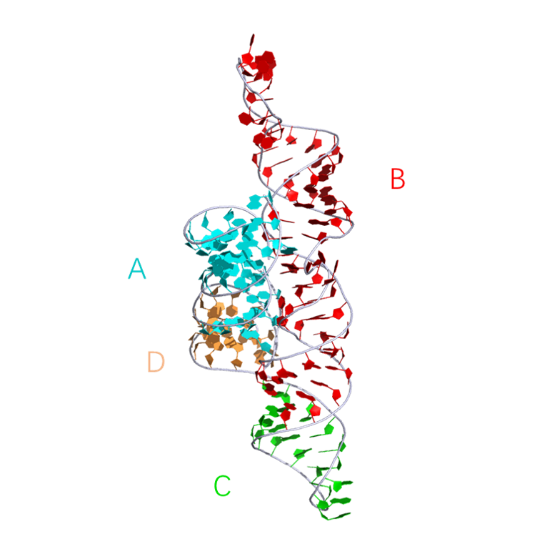
3D visualisation
The structure and active center of the hairpin ribozyme in its natural four-way junction form. This representation was generated from PDB ID: 1M5O at 2.2 Å resolution.
 |
|
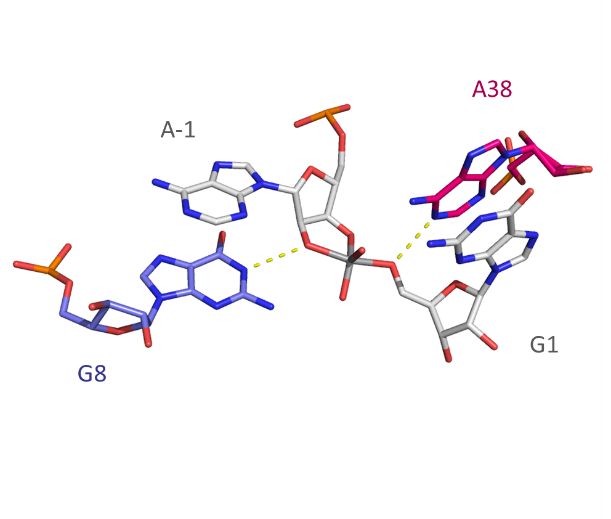
Catalytic centre
The active site, with a vanadate replacing the scissile phosphate as a transition state analog. The geometry of the vanadium dictates that this structure approximates to that of a phosphorane with inline attack. G8 N1 and A38 N1 are hydrogen bonded to the O2′ and O5′ , respectively, shown by AQ4 broken lines.(left)
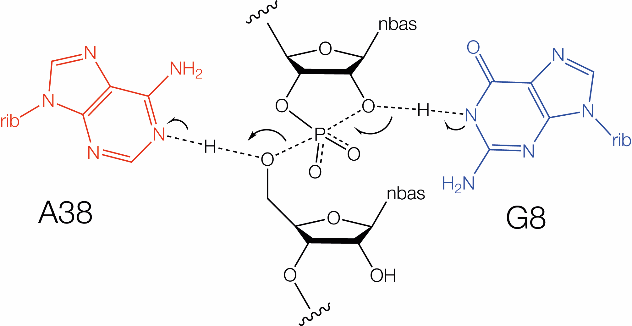
The hairpin ribozyme and a proposed reaction mechanism. Proposed mechanism of cleavage and ligation based on general acid-base catalysis. This depicts the transition state; the arrows show the flow of electrons for cleavage.(right)
 |
 |
Chemical mechanism
The hairpin ribozyme employs general acid-base catalysis, using the nucleobases of G8 (in its deprotonated form) as general base to remove the proton from the 2'-OH nucleophile and A38 (in its protonated form) as the general acid to protonate the leaving group. The ribozyme (together with the VS ribozyme) is one of the standard G+A mechanism ribozymes. In the crystal structure of a transition state analog [5] G8 N1 interacts with the O2' nucleophile and A38 N1 with the O5' leaving group as expected from the mechanism of cleavage. The rates of cleavage and ligation of both ribozymes are strongly pH dependent[6,16], consistent with proton transfer occurring in the transition state. Reaction rates increase at low pH, plateau around neutrality, and remain up to pH 9. No reduction in activity was found at the high pH end, i.e. no there is no bell-shaped pH dependence similar to that of the VS ribozyme. However, the upper pKa would not be detectable if ≥ 10. However, a bell-shaped pH dependence of reaction rate resulted when G8 was substituted with nucleobases of lower pKa such as 2,6-diaminopurine with a pKa around 5.4 [6,10] and imidazole with a pKa around neutrality [17]. The assignment of G8 as general base and A38 as general acid in the cleavage reaction were confirmed by phosphorothiolate substitution experiments [10].
References
[1] Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA.
Buzayan, J. M., W. L. Gerlach and G. Bruening
Nature.(1986)
[2] Essential nucleotide sequences and secondary structure elements of the hairpin ribozyme.
Berzal-Herranz, A., S. Joseph, B. Chowrira, S. Butcher and J. Burke
The EMBO journal 12(6): 2567-2573.(1993)
[3] A unique mechanism for RNA catalysis: the role of metal cofactors in hairpin ribozyme cleavage.
Hampel, A. and J. Cowan
Chemistry & biology 4(7): 513-517.(1997)
[4] Mutational analysis of loops 1 and 5 of the hairpin ribozyme.
Shippy, R., A. Siwkowski and A. Hampel
Biochemistry 37(2): 564-570.(1998)
[5] Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis.
Rupert, P. and A. Ferré-D'Amaré
Nature 410(6830): 780-786.(2001)
[6] Functional involvement of G8 in the hairpin ribozyme cleavage mechanism.
Pinard, R., K. Hampel, J. Heckman, D. Lambert, P. Chan, F. Major and J. Burke
The EMBO journal 20(22): 6434-6442.(2001)
[7] Transition state stabilization by a catalytic RNA.
Rupert, P., A. Massey, S. Sigurdsson and A. Ferré-D'Amaré
Science (New York, N.Y.) 298(5597): 1421-1424.(2002)
[8] Role of an active site adenine in hairpin ribozyme catalysis.
Kuzmin, Y., C. Da Costa, J. Cottrell and M. Fedor
Journal of molecular biology 349(5): 989-1010.(2005)
[9] Water in the active site of an all-RNA hairpin ribozyme and effects of Gua8 base variants on the geometry of phosphoryl transfer.
Salter, J., J. Krucinska, S. Alam, V. Grum-Tokars and J. Wedekind
Biochemistry 45(3): 686-700.(2006)
[10] General acid-base catalysis mediated by nucleobases in the hairpin ribozyme.
Kath-Schorr, S., T. Wilson, N. Li, J. Lu, J. Piccirilli and D. Lilley
Journal of the American Chemical Society 134(40): 16717-16724.(2012)
[11] Engineering of hairpin ribozyme variants for RNA recombination and splicing.
Hieronymus, R. and S. Müller
Annals of the New York Academy of Sciences 1447(1): 135-143.(2019)
[12] Prebiotically Plausible RNA Activation Compatible with Ribozyme-Catalyzed Ligation.
Song, E., E. Jiménez, H. Lin, K. Le Vay, R. Krishnamurthy and H. Mutschler
Angewandte Chemie (International ed. in English) 60(6): 2952-2957.(2021)
[13] Identification of over 200-fold more hairpin ribozymes than previously known in diverse circular RNAs.
Weinberg, C., V. Olzog, I. Eckert and Z. Weinberg
Nucleic acids research 49(11): 6375-6388.(2021)
[14] ViroidDB: a database of viroids and viroid-like circular RNAs.
Lee, B., U. Neri, C. Oh, P. Simmonds and E. Koonin
Nucleic acids research 50: D432-D438.(2022)
[15] RNA self-splicing by engineered hairpin ribozyme variants.
Hieronymus, R., J. Zhu and S. Müller
Nucleic acids research 50(1): 368-377.(2022)
[16] Observation of internal cleavage and ligation reactions of a ribozyme.
Nahas, M.K., Wilson, T.J., Hohng, S., Jarvie, K., Lilley, D.M.J. and Ha, T.
Nature Struct. Molec. Biol., 11, 1107-1113.(2004)
[17] Nucleobase catalysis in the hairpin ribozyme.
Wilson, T.J., Ouellet, J., Zhao, Z.Y., Harusawa, S., Araki, L., Kurihara, T. and Lilley, D.M.J.
RNA, 12, 980-987.(2006)
 Home
Home Database
Database Research
Research About us
About us

