Hatchet
Rfam ID: RF02678
click into different sections:
Timeline
-
2015 Discovery, Secondary structure[1]
-
2015 Biochemical analysis of hatchet ribozyme[2]
-
2018 SHAPE probing of pre-catalytic folds of hatchet ribozyme[3]
-
2019 Crystal structure and cleavage mechanism[4]
Description
The hatchet ribozyme is an RNA structure that catalyzes its own cleavage at a specific site. In other words, it is a self-cleaving ribozyme. Hatchet ribozymes were discovered by a bioinformatics strategy as RNAs Associated with Genes Associated with Twister and Hammerhead ribozymes, or RAGATH. The consensus sequence and secondary structure of this class includes 13 highly conserved and numerous other modestly conserved nucleotides inter-dispersed among bulges linking four base-paired substructures. A representative hatchet ribozyme requires divalent cations such as Mg2+ to promote RNA strand scission with a maximum rate constant of ~4/min. As with all other small self-cleaving ribozymes discovered to date, hatchet ribozymes employ a general mechanism for catalysis consisting of a nucleophilic attack of a ribose 2′-oxygen atom on the adjacent phosphorus center. Kinetic characteristics of the reaction demonstrate that members of this ribozyme class have an essential requirement for divalent metal cations and that they have a complex active site which employs multiple catalytic strategies to accelerate RNA cleavage by internal phosphoester transfer.
Structure and mechanism
2D representation
Secondary structure of the Hatchet ribozyme. The sequence is color coded according to helical segments observed in the tertiary structure.

|
3D visualisation
Crystal structure of the Hatchet ribozyme. This representation was generated from PDB ID: 6JQ5 at 2.06 Å resolution. Helices are differentiated by color.
 |
|
Catalytic centre
The proposed model of the cleavage site of the Hatchet ribozyme. The modeled in-line alignment conformation indicates the potential nucleotides that may contribute to general base and general acid catalysis in the cleavage process.
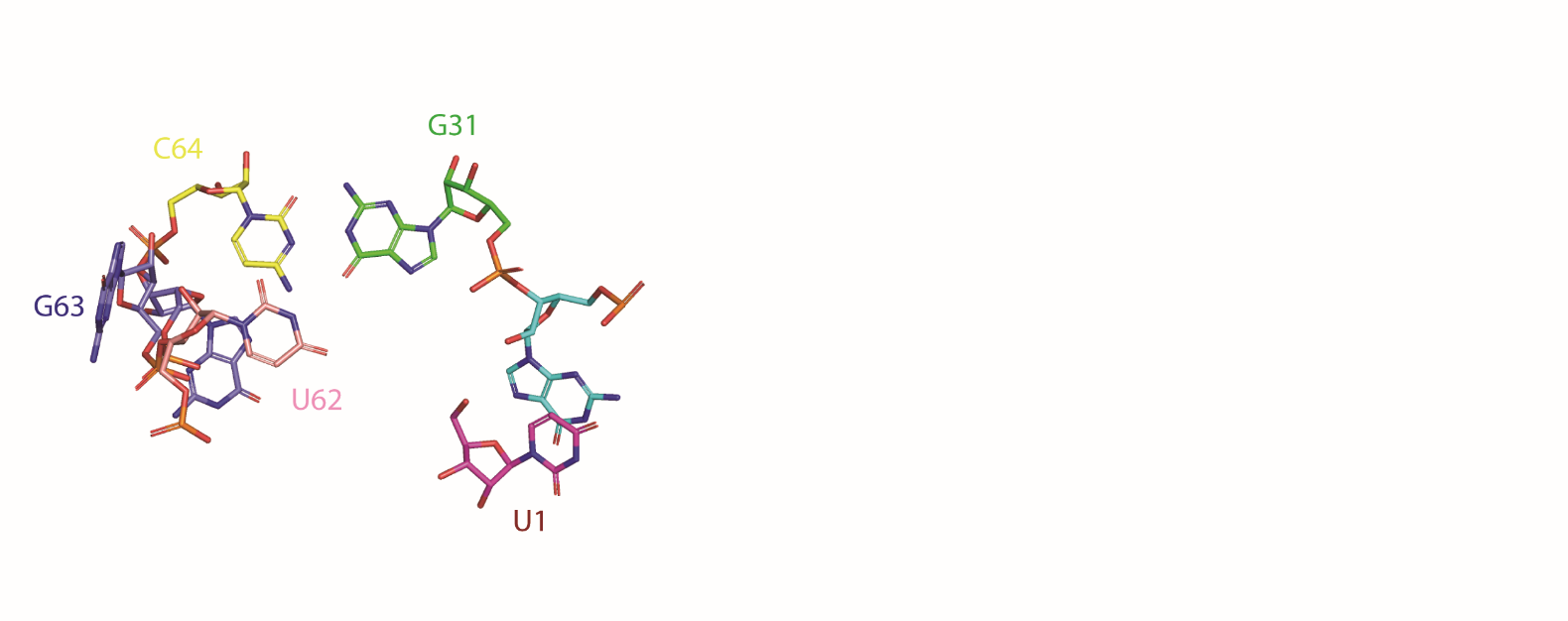 |
References
[1] New classes of self-cleaving ribozymes revealed by comparative genomics analysis.
Weinberg, Z., P. B. Kim, T. H. Chen, S. Li, K. A. Harris, C. E. Lunse and R. R. Breaker
Nat Chem Biol 11(8): 606-10.(2015)
[2] Biochemical analysis of hatchet self-cleaving ribozymes.
Li, S., C. E. Lunse, K. A. Harris and R. R. Breaker
RNA 21(11): 1845-51.(2015)
[3] SHAPE probing pictures Mg2+-dependent folding of small self-cleaving ribozymes.
Gasser, C., J. Gebetsberger, M. Gebetsberger and R. Micura
Nucleic Acids Res 46(14): 6983-6995.(2018)
[4] Hatchet ribozyme structure and implications for cleavage mechanism.
Zheng, L., C. Falschlunger, K. Huang, E. Mairhofer, S. Yuan, J. Wang, D. J. Patel, R. Micura and A. Ren
Proc Natl Acad Sci U S A 116(22): 10783-10791.(2018)
[5] Fundamental studies of functional nucleic acids: aptamers, riboswitches, ribozymes and DNAzymes.
Micura, R. and C. Hobartner
Chem Soc Rev 49(20): 7331-7353.(2020)
 Home
Home Database
Database Research
Research About us
About us

