Spliceosome
click into different sections:
Timeline
-
1977 Splicing phenomenon found[1]
-
1983 First isolation of spliceosome subunits[2]
-
1985 In vitro splicing experiment[3]
-
1985 In vitro splicing experiment[4]
-
1992 Demonstrate a conserved base-pairing interaction between the U6 and U2 snRNAs that is mutually exclusive with the U4-U6 interaction[5]
-
1993 Proposed that the two phosphotransesterifications of splicing are catalyzed by a two-metal mechanism[6]
-
1997 Divalent metals stabilize the leaving group during each step of splicing[7]
-
2000 Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome[8]
-
2001 U2 and U6 can base-pair and fold in vitro into a structure that catalyzes reactions similar to the two steps of pre-mRNA splicing[9]
-
2009 U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps[10]
-
2013 Provides crucial insights into the architecture of the spliceosome active site, and reinforces the notion that nuclear pre-mRNA splicing and group II intron splicing have a common origin.[11]
-
.
2013 Demonstrate that RNA mediates catalysis within the spliceosome[12]
-
2015 S.p ILS, 3.6 Å & The first atomic structure of the intact spliceosome[13]
-
2016 The resolution of tri-snRNP, a complex during splice assembly, was increased to 5.9 angstroms[14]
-
2016 S.c C, 3.4 Å & Active site after branching[15]
-
2016 U4/U6.U5 tri-snRNP,3.8Å[16]
-
2016 B act , 3.5 Å & Catalytic center is formed[17]
-
2017 S.c C*, 4.0 Å[18]
-
2017 S.c C*, 3.8 Å[19]
-
2017 S.c B, 7.2 (3.7) Å[20]
-
2017 S.c ILS, 3.5 Å[21]
-
2017 S.c P, 3.3 Å[22]
-
2017 S.c P, 3.6 Å[23]
-
2017 C*, 5.9 Å[24]
-
2017 C*, 3.8 ÅThe first atomic model of human Spliceosome[25]
-
2017 B, 9.9 (4.5) Å[26]
-
2018 S.c pre-B, 3.3-4.6 Å & S.c B, 3.9 Å[27]
-
2018 S.c A, 4.9 (4.0) Å[28]
-
2018 C, 4.1 Å[29]
-
2018 Bact , 3.4 Å (core)[30]
-
2018 pre-B (5.7 Å) and B (3.8 Å)[31]
-
2019 S.c B*, 2.9-3.8 ÅFour distinct structures on two different substrates[32]
-
2019 P, 3.3 Å[33]
-
2019 P (3.0 Å) and ILS (2.9 Å)[34]
-
2019 pre-B, 3.3 Å Mechanism of 5' splice site transfer for human spliceosome activation[35]
-
2019 Mechanism of Branching[36]
-
2020 The structure of 17s U2 snRNP was analyzed and a complete molecular model of 17s U2 snRNP was obtained[37]
-
2020 Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation[38]
-
2021 Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2[39]
-
2022 A series of high-resolution (2.0-2.2 Å) U2 snRNP structures were identified[40]
Description
The spliceosome is a highly dynamic and heterogeneous metal ribozyme; During the splicing reaction, the dynamic spliceosome has an immobile core of about 20 protein and RNA components, which are organized around a conserved splicing active site. The divalent metal ions, coordinated by U6 small nuclear RNA (snRNA), catalyze the branching reaction and exon ligation.
Choreography of the RNA elements during the splicing cycle
Choreography of the RNA elements during the splicing cycle. Figure based on [42]. The spliceosomal assembly begins with recognition of 5'SS, BPS, and 3'SS. The U1 snRNP and other splicing factors associate with pre-mRNA strand to assemble the E complex. The U2 snRNA base pairs with BPS to yield a pre-spliceosomal A complex, which then binds the U4/U6.U5 tri-snRNP to form the pre-B complex. Pre-B complex to B complex is activated by prp28, during which U1 snRNP dissociates and 5 'SS translocates to the vicinity of U6 snRNA. Under the activation of brr2, U4 snRNP dissociates and forms B act complex. In the B act complex,the 5'SS and 5'-exons are recognized by the ACAGA box of U6 snRNA and the loop I of U5 snRNA, respectively. The B act complex is converted to B * by the action of Prp2. Catalysis begins in the resulting B *complex and branching begins with the addition of step I factors. Branching reaction to occur at the 5'SS, producing the free 5'-exon and the intron lariat structure[44]. The C complex is reshaped by Prp16 to move the lariat junction away from the active site and to allow proper engagement of the 3'SS. In the C *complex, exon ligation occurs in the presence of step II. Driven by Prp22, the spliceosome begins to disintegrate and the ligated exon is released from the P complex. The color scheme is as follows: U1 (yellow), U2 (green), U4 (orange), U5 (pink), U6 (blue) snRNAs; intron of pre-mRNA (magenta); and two exons of pre-mRNA : 5'-exton(red),3'-exton (orange). Abbreviations: BPS, branch point sequence; pre-mRNA, precursor messenger RNA; SS, splice site; ILS, intron lariat spliceosome.

Transesterification Reactions
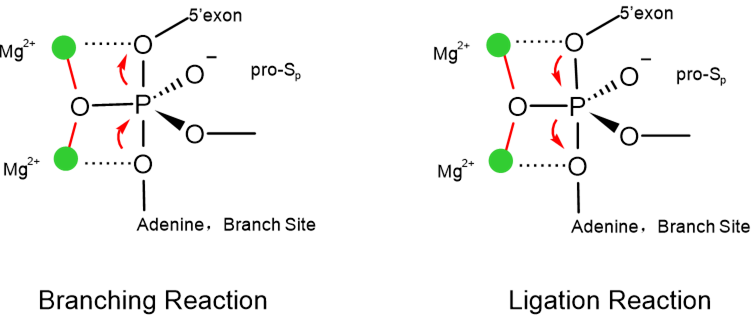
The transesterification Reactions in the Spliceosome. The branching and ligation reactions require formation of a trigonal planar transition state that is stabilized by two magnesium ions. Figure based on [12]. The two transesterification reactions occur at the same active site in the spliceosome. Magnesium ions are required in the active site for catalysis, and they function to stabilize the transition states. The positive charge on the magnesium ion interacts with the negatively charged oxygen leaving group. This interaction reduces oxygen's attraction to the positively charged phosphate, which allows it to dissociate more readily. In both the catalytic reactions, magnesium ions interact with nucleotides in U6 snRNA and the pre-mRNA strand. The snRNA and pre-mRNA interact with the catalytic metal ion through the oxygen attached to the phosphate backbone. There are two non-bridging oxygens attached to each phosphate.
References
[1] An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA.
Chow, L. T., R. E. Gelinas, T. R. Broker and R. J. Roberts
Cell 12(1): 1-8.(1977)
[2] The U1 small nuclear RNA-protein complex selectively binds a 5' splice site in vitro.
Mount, S. M., I. Pettersson, M. Hinterberger, A. Karmas and J. A. Steitz
Cell 33(2): 509-518.(1983)
[3] Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences.
Frendewey, D. and W. Keller
Cell 42(1): 355-367.(1985)
[4] A multicomponent complex is involved in the splicing of messenger RNA precursors.
Grabowski, P., S. Seiler and P. Sharp
Cell 42(1): 345-353.(1985)
[5] A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome.
Madhani, H. D. and C. Guthrie
Cell 71(5): 803-817.(1992)
[6] A general two-metal-ion mechanism for catalytic RNA.
Steitz, T. A. and J. A. Steitz
Proc Natl Acad Sci U S A 90(14): 6498-6502.(1993)
[7] Metal ion catalysis during splicing of premessenger RNA.
Sontheimer, E. J., S. Sun and J. A. Piccirilli
Nature 388(6644): 801-805.(1997)
[8] Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome.
Yean, S. L., G. Wuenschell, J. Termini and R. J. Lin
Nature 408(6814): 881-884.(2000)
[9] Splicing-related catalysis by protein-free snRNAs.
Valadkhan, S. and J. L. Manley
Nature 413(6857): 701-707.(2001)
[10] Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps.
Mefford, M. A. and J. P. Staley
RNA 15(7): 1386-1397.(2009)
[11] Crystal structure of Prp8 reveals active site cavity of the spliceosome.
Galej, W. P., C. Oubridge, A. J. Newman and K. Nagai
Nature 493(7434): 638-643.(2013)
[12] RNA catalyses nuclear pre-mRNA splicing.
Fica, S. M., N. Tuttle, T. Novak, N. S. Li, J. Lu, P. Koodathingal, Q. Dai, J. P. Staley and J. A. Piccirilli
Nature 503(7475): 229-234.(2013)
[13] Structure of a yeast spliceosome at 3.6-angstrom resolution.
Yan, C., J. Hang, R. Wan, M. Huang, C. C. Wong and Y. Shi
Science 349(6253): 1182-1191.(2015)
[14] Cryo-EM structure of the spliceosome immediately after branching.
Galej, W. P., M. E. Wilkinson, S. M. Fica, C. Oubridge, A. J. Newman and K. Nagai
Nature 537(7619): 197-201.(2016)
[15] Structure of a yeast catalytic step I spliceosome at 3.4 A resolution.
Wan, R., C. Yan, R. Bai, G. Huang and Y. Shi
Science 353(6302): 895-904.(2016)
[16] The 3.8 A structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis.
Wan, R., C. Yan, R. Bai, L. Wang, M. Huang, C. C. Wong and Y. Shi
Science 351(6272): 466-475.(2016)
[17] Structure of a yeast activated spliceosome at 3.5 A resolution.
Yan, C., R. Wan, R. Bai, G. Huang and Y. Shi
Science 353(6302): 904-911.(2016)
[18] Structure of a yeast step II catalytically activated spliceosome.
Yan, C., R. Wan, R. Bai, G. Huang and Y. Shi
Science 355(6321): 149-155.(2017)
[19] Structure of a spliceosome remodelled for exon ligation.
Fica, S. M., C. Oubridge, W. P. Galej, M. E. Wilkinson, X. C. Bai, A. J. Newman and K. Nagai
Nature 542(7641): 377-380.(2017)
[20] Structure of a pre-catalytic spliceosome.
Plaschka, C., P. C. Lin and K. Nagai
Nature 546(7660): 617-621.(2017)
[21] Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae.
Wan, R., C. Yan, R. Bai, J. Lei and Y. Shi
Cell 171(1): 120-132 e112.(2017)
[22] Structure of the yeast spliceosomal postcatalytic P complex.
Liu, S., X. Li, L. Zhang, J. Jiang, R. C. Hill, Y. Cui, K. C. Hansen, Z. H. Zhou and R. Zhao
Science 358(6368): 1278-1283.(2017)
[23] Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae.
Bai, R., C. Yan, R. Wan, J. Lei and Y. Shi
Cell 171(7): 1589-1598 e1588.(2017)
[24] Cryo-EM Structure of a Pre-catalytic Human Spliceosome Primed for Activation.
Bertram, K., D. E. Agafonov, O. Dybkov, D. Haselbach, M. N. Leelaram, C. L. Will, H. Urlaub, B. Kastner, R. Luhrmann and H. Stark
Cell 170(4): 701-713 e711.(2017)
[25] An Atomic Structure of the Human Spliceosome.
Zhang, X., C. Yan, J. Hang, L. I. Finci, J. Lei and Y. Shi
Cell 169(5): 918-929 e914.(2017)
[26] Cryo-EM structure of a human spliceosome activated for step 2 of splicing.
Bertram, K., D. E. Agafonov, W. T. Liu, O. Dybkov, C. L. Will, K. Hartmuth, H. Urlaub, B. Kastner, H. Stark and R. Luhrmann
Nature 542(7641): 318-323.(2017)
[27]Structures of the fully assembled Saccharomyces cerevisiae spliceosome before activation.
Bai, R., R. Wan, C. Yan, J. Lei and Y. Shi
Science 360(6396): 1423-1429.(2018)
[28] Prespliceosome structure provides insights into spliceosome assembly and regulation.
Plaschka, C., P. C. Lin, C. Charenton and K. Nagai
Nature 559(7714): 419-422.(2018)
[29] Structure of a human catalytic step I spliceosome.
Zhan, X., C. Yan, X. Zhang, J. Lei and Y. Shi
Science 359(6375): 537-545.(2018)
[30] Structure and Conformational Dynamics of the Human Spliceosomal B(act) Complex.
Haselbach, D., I. Komarov, D. E. Agafonov, K. Hartmuth, B. Graf, O. Dybkov, H. Urlaub, B. Kastner, R. Luhrmann and H. Stark
Cell 172(3): 454-464 e411.(2018)
[31] Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Zhan, X., C. Yan, X. Zhang, J. Lei and Y. Shi
Cell Res 28(12): 1129-1140.(2018)
[32] Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Wan, R., R. Bai, C. Yan, J. Lei and Y. Shi
Cell 177(2): 339-351 e313.(2019)
[33] A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation.
Fica, S. M., C. Oubridge, M. E. Wilkinson, A. J. Newman and K. Nagai
Science 363(6428): 710-714(2019)
[34] Structures of the human spliceosomes before and after release of the ligated exon.
Zhang, X., X. Zhan, C. Yan, W. Zhang, D. Liu, J. Lei and Y. Shi
Cell Res 29(4): 274-285.(2019)
[35] Mechanism of 5' splice site transfer for human spliceosome activation.
Charenton, C., M. E. Wilkinson and K. Nagai
Science 364(6438): 362-367.
[36] Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Wan, R., R. Bai, C. Yan, J. Lei and Y. Shi
Cell 177(2): 339-351 e313.(2019)
[37] Molecular architecture of the human 17S U2 snRNP.
Zhang, Z., C. L. Will, K. Bertram, O. Dybkov, K. Hartmuth, D. E. Agafonov, R. Hofele, H. Urlaub, B. Kastner, R. Luhrmann and H. Stark
Nature 583(7815): 310-313.(2020)
[38] Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Townsend, C., M. N. Leelaram, D. E. Agafonov, O. Dybkov, C. L. Will, K. Bertram, H. Urlaub, B. Kastner, H. Stark and R. Luhrmann
Science 370(6523).(2020)
[39] Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Bai, R., R. Wan, C. Yan, Q. Jia, J. Lei and Y. Shi
Science 371(6525).(2021)
[40] Structural basis of branch site recognition by the human spliceosome.
Tholen, J., M. Razew, F. Weis and W. P. Galej
Science 375(6576): 50-57.(2022)
[41] Molecular Mechanisms of pre-mRNA Splicing through Structural Biology of the Spliceosome.
Yan, C., R. Wan and Y. Shi
Cold Spring Harb Perspect Biol 11(1).(2019)
[42] How Is Precursor Messenger RNA Spliced by the Spliceosome?
Wan, R., R. Bai, X. Zhan and Y. Shi
Annu Rev Biochem 89: 333-358.(2020)
[43] RNA Splicing by the Spliceosome.
Wilkinson, M. E., C. Charenton and K. Nagai
Annu Rev Biochem 89: 359-388.(2020)
[44] Mechanistic insights into precursor messenger RNA splicing by the spliceosome.
Shi, Y.
Nat Rev Mol Cell Biol, 18, 655-670.(2017)
 Home
Home Database
Database Research
Research About us
About us

