GlmS
Rfam ID: RF00234
click into different sections:
Timeline
-
2004 Discovery,Secondary structure[1]
-
2006 Pseudoknot structure[2]
-
2006 Crystal structure[3]
-
2007 Catalytic mechanism[4]
-
2007 Mechanism of mRNA destabilization by the GlmS ribozyme[6]
-
2009 Chemical Mechanism[7]
-
2010 Use of a small molecule coenzyme by a gene-regulatory RNA[8]
-
2010 The role of Mg2+ in active sites[9]
-
2011 The GlmS riboswitch integrates signals from activating and inhibitory metabolites in vivo[10]
-
2011 An expanded collection and refined consensus model of GlmS ribozymes[11]
-
2012 The GlmS ribozyme cofactor is a general acid-base catalyst[12]
-
2013 An in vitro evolved GlmS ribozyme has the wild-type fold but loses coenzyme dependence[13]
-
2017 GlcN6P cofactor play a variety of catalytic roles in GlmS ribozyme[15]
-
2018 The GlmS ribozyme could be used as a tool to study essential genes in T. brucei[16]
-
2020 Comprehensive sequence-to-function mapping of cofactor-dependent RNA catalysis in the GlmS ribozyme[17]
-
2021 The GlmS ribozyme is a very suitable target for antibacterial drug development with antisense oligonucleotides[18]
Description
The hairpin ribozyme and substrate originate from satellite RNAs associated with three different plant RNA viruses . These are the negative strands of the satellite RNAs from tobacco ringspot virus (sTRSV), chicory yellow mottle virus type 1 (sCYMV1), and arabis mosaic virus (sArMV). These satellite RNAs are singlestranded RNA molecules that replicate via a rolling circle mechanism. The catalytic center of this satellite RNA was determined, reducing the number of nucleotides necessary for catalysis to a 50nt ribozyme cutting a 14nt substrate in a trans reaction. The overall secondary structure of this catalytic center was hairpin-like and was therefore named the hairpin ribozyme.
Structure and mechanism
Secondary structure depictions of the GlmS ribozyme with the scissile phosphates (blue) arrowed and the base (blue) and cofactor Glc6P(red) are shown.2D representation

|
The structure of the GlmS ribozyme bound to glucose-6-phosphate. This representation was generated from PDB ID: 2HO7 at 2.9 Å resolution.3D visualisation
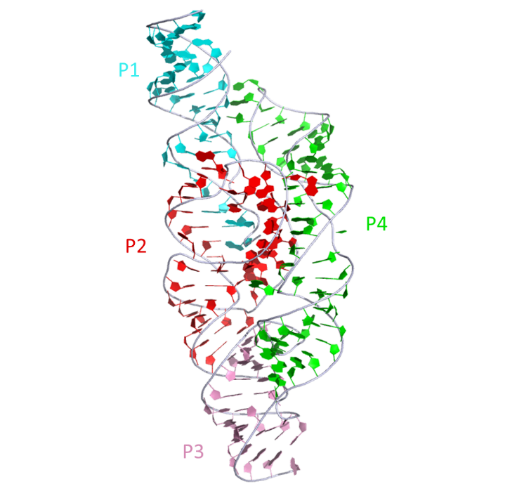 |
|
Catalytic centre
Close view of the catalytic center of the ribozyme. The A-0 O2' is positioned for inline nucleophilic attack (broken red line). G40 N1 is well placed to act as general base. An amino group has been modeled onto Glc6P to generate glucosamine-6-phosphate; this amine is positioned to act as general acid to protonate the G-1 O5' -leaving group.
The proposed catalytic mechanism of the GlmS ribozyme. In the nucleolytic cleavage reaction the 2'-OH of a ribose acts as the nucleophile, attacking the scissile phosphate and releasing the 5'-oxygen leaving group. This reaction requires an in-line (180°) conformation for the 2'-OH, scissile phosphate and 5'-oxygen. In the GlmS ribozyme, a guanosine has been implicated in the reaction mechanism.
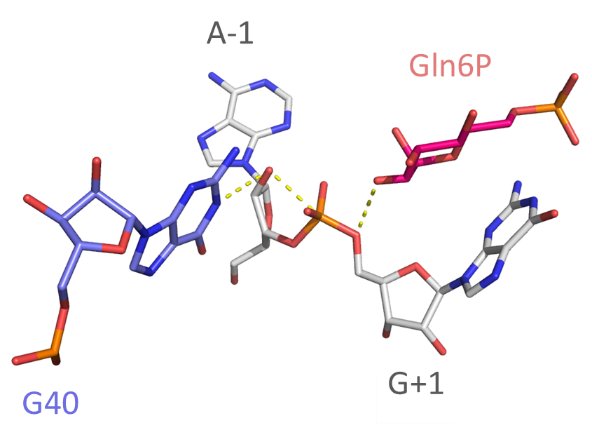 |
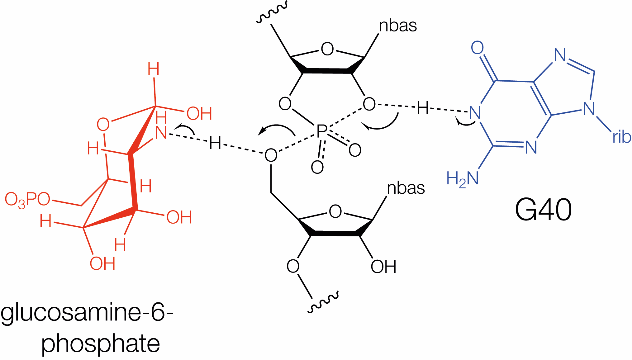 |
Chemical mechanism
Uniquely GlmS is a ribozyme that uses a co-enzyme, and is both a riboswitch and a ribozyme. GlmS exploits its glucosamine-6-phosphate (Glc6P) ligand as a participant in the chemistry of the catalysis of phosphoryl transfer. The riboswitch was found to undergo self-cleavage, creating 5'-OH and cyclic 2'3'-termini [1]. The activity required the presence of Glc6P, with half-maximal activity at ~200 μM. GlmS was found to be active in the presence of a number of divalent metal ions, and also active in high concentrations of monovalent metal ions and Co(III) hexammine [19], indicating that no direct involvement of a metal ion is likely. In the crystal [3,4,7] the amine of the Glc6P is 2.9Å from the O5' leaving group, so well positioned to act as general acid. The pKa of the amine is 8.2 and so Glc6P should readily donate a proton to the O5’ oxyanion. G40 N1 (in B. subtilis; the corresponding nucleotide in B. anthracis is G33) is 3.2 Å from the O2' nucleophile, suggesting a role as general base. A GlmS G40A mutant ribozyme was severely impaired in cleavage, with a rate of kobs = 5 x 10-5 min-1 [5]. The rate of cleavage was found to depend on pH, but because the binding of the ligand is pH dependent, this is difficult to interpret unfortunately. However the probable mechanism of the GlmS ribozyme is general acid-base catalysis, with a guanine N1 as general base and the amine of the exogenous Glc6P ligand acting as general acid.
References
[1] Control of gene expression by a natural metabolite-responsive ribozyme
Winkler, W., A. Nahvi, A. Roth, J. Collins and R. Breaker
Nature 428(6980): 281-286.(2004)
[2] Core requirements for glmS ribozyme self-cleavage reveal a putative pseudoknot structure.
Soukup, G.
Nucleic acids research 34(3): 968-975.(2006).
[3] Structural basis of glmS ribozyme activation by glucosamine-6-phosphate.
Klein, D. and A. Ferré-D'Amaré
Science (New York, N.Y.) 313(5794): 1752-1756.(2006)
[4] Structural investigation of the GlmS ribozyme bound to Its catalytic cofactor.
Cochrane, J., S. Lipchock and S. Strobel
Chemistry & biology 14(1): 97-105.(2007)
[5] Essential role of an active-site guanine in glmS ribozyme catalysis.
Klein, D., M. Been and A. Ferré-D'Amaré
Journal of the American Chemical Society 129(48): 14858-14859.(2007)
[6] Mechanism of mRNA destabilization by the glmS ribozyme.
Collins, J., I. Irnov, S. Baker and W. Winkler
Genes & development 21(24): 3356-3368.(2007)
[7] Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme.
Cochrane, J., S. Lipchock, K. Smith and S. Strobel
Biochemistry 48(15): 3239-3246.(2009)
[8] The glmS ribozyme: use of a small molecule coenzyme by a gene-regulatory RNA.
Ferré-D'Amaré, A.
Quarterly reviews of biophysics 43(4): 423-447.(2010)
[9] Analysis of metal ion dependence in glmS ribozyme self-cleavage and coenzyme binding.
Klawuhn, K., J. Jansen, J. Souchek, G. Soukup and J. Soukup
Chembiochem : a European journal of chemical biology 11(18): 2567-2571.(2010)
[10] The glmS riboswitch integrates signals from activating and inhibitory metabolites in vivo.
Watson, P. and M. Fedor
Nature structural & molecular biology 18(3): 359-363.(2011)
[11] An expanded collection and refined consensus model of glmS ribozymes.
McCown, P., A. Roth and R. Breaker
RNA (New York, N.Y.) 17(4): 728-736.(2011)
[12] The glmS ribozyme cofactor is a general acid-base catalyst.
Viladoms, J. and M. Fedor
Journal of the American Chemical Society 134(46): 19043-19049.(2012)
[13] An in vitro evolved glmS ribozyme has the wild-type fold but loses coenzyme dependence.
Lau, M. W. L. and A. R. Ferré-D Amaré
Nature Chemical Biology 9(12): 805.(2013)
[14] Activation of the glmS Ribozyme Confers Bacterial Growth Inhibition.
Schüller, A., D. Matzner, C. Lünse, V. Wittmann, C. Schumacher, S. Unsleber, H. Brötz-Oesterhelt, C. Mayer, G. Bierbaum and G. Mayer
Chembiochem : a European journal of chemical biology 18(5): 435-440.(2017)
[15] The GlcN6P cofactor plays multiple catalytic roles in the glmS ribozyme.
Bingaman, J., S. Zhang, D. Stevens, N. Yennawar, S. Hammes-Schiffer and P. Bevilacqua
Nature chemical biology 13(4): 439-445.(2017)
[16] A Riboswitch-based Inducible Gene Expression System for Trypanosoma brucei.
Cruz-Bustos, T., S. Ramakrishnan, C. Cordeiro, M. Ahmed and R. Docampo
The Journal of eukaryotic microbiology 65(3): 412-421.(2018)
[17] Comprehensive sequence-to-function mapping of cofactor-dependent RNA catalysis in the glmS ribozyme.
Andreasson, J., A. Savinov, S. Block and W. Greenleaf
Nature communications 11(1): 1663.(2020)
[18] Targeting glmS Ribozyme with Chimeric Antisense Oligonucleotides for Antibacterial Drug Development.
Traykovska, M., K. Popova and R. Penchovsky
ACS synthetic biology 10(11): 3167-3176.(2021)
[19] Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions.
Roth, A., Nahvi, A., Lee, M., Jona, I. and Breaker, R.R.
RNA, 12, 607-619(2006)
 Home
Home Database
Database Research
Research About us
About us

