Group I self-splicing intron
Rfam ID: RF00028
click into different sections:
Timeline
-
1982 Discovery[1]
-
1982 Determination of shared secondary structure[2]
-
1986 The intervening sequence RNA of Tetrahymena is an enzyme[3]
-
1988 ωG is closely related to the choice of 3' splice site[4]
-
1990 3D models of group I intron based on comparative sequence analysis[8]
-
1996 Crystal structure of Tetrahymena P4-P6 domain[9]
-
1998 Crystal structure of an engineered, active Tetrahymena ribozyme at 5.0 Å resolution[10]
-
2004 Crystal structure of Azoarcus group I intron with both exons[11]
-
2004 Crystal structure of an active Tetrahymena ribozyme[12]
-
2005 Crystal structure of phage Twort group I ribozyme-product complex[13]
-
2005 Crystal structure of a catalytically active Azoarcus group I intron splicing intermediate[14]
-
2008 Sequence and structure database[15]
-
2011 Long-range tertiary contacts in RNA exhibit distinct catalytic roles[16]
-
2021 Cryo-EM structures of full-length Tetrahymena ribozyme[17]
-
2022 Tetrahymena group I intron at 2.98-Å resolution overall (2.85 Å for the core)[18]
Description
Group I introns are large self-splicing ribozymes. They catalyze their own excision from mRNA, tRNA and rRNA precursors in a wide range of organisms. The core secondary structure consists of nine paired regions (P1-P9). These fold to essentially two domains—the P4-P6 domain (formed from the stacking of P5, P4, P6 and P6a helices) and the P3-P9 domain (formed from the P8, P3, P7 and P9 helices). The secondary structure mark-up for this family represents only this conserved core. Group I introns often have long open reading frames inserted in loop regions
Structure and mechanism
Secondary structure of the apo L-21 ScaI Tetrahymena ribozyme based on the cryo-EM model, a linear form of the self-splicing intron without its first 21 nucleotides (nts 22–409)2D representation

|
Cryo-EM structures of full-length Tetrahymena ribozyme. The overall structure of the Apo L-21 ScaI Tetrahymena ribozyme was generated from PDB ID:7EZ0 at 3.1 Å resolution.3D visualisation
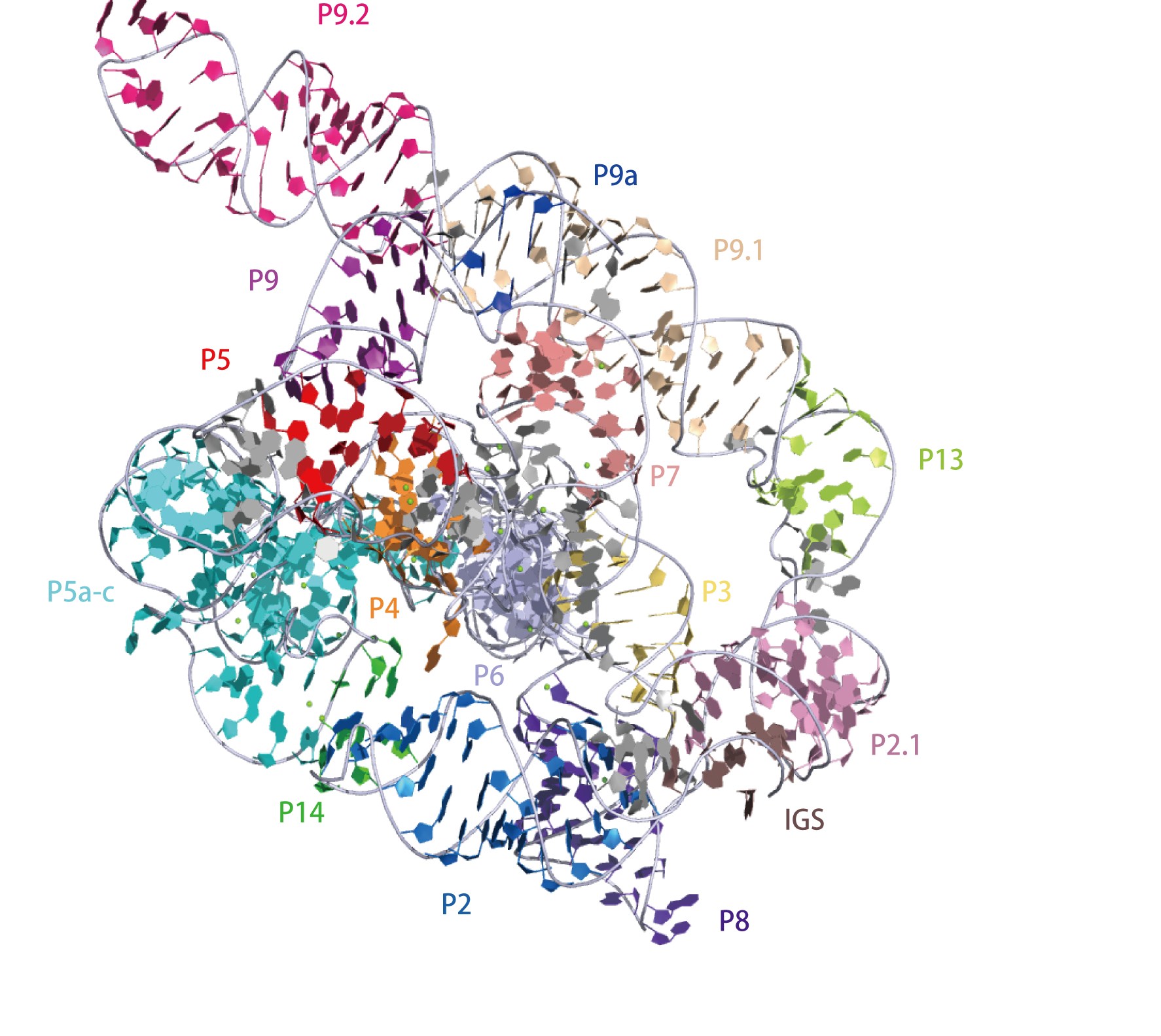 |
|
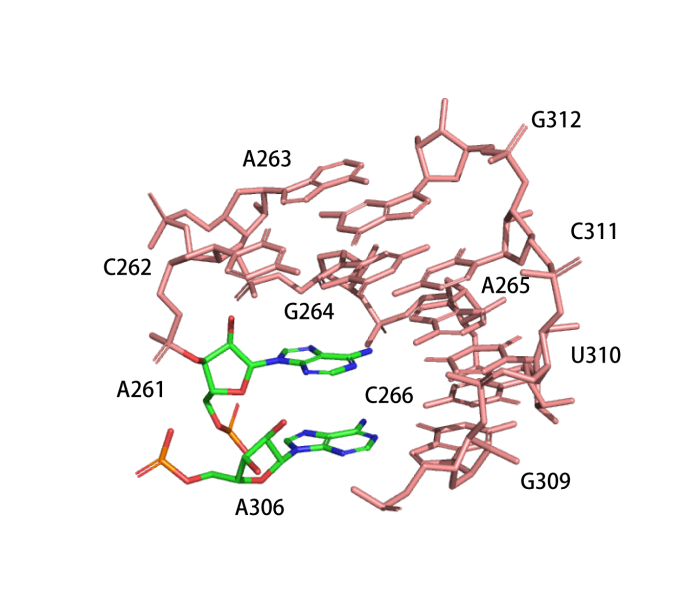
Catalytic centre
The active centre of the ribozyme is composed of four layers of base triples.(PDB ID: 7EZ0)
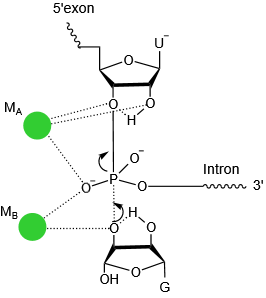
The transition state of first step catalysis.MB binds to the 3’-hydroxyl group of the exogenous guanosine in the first step, functioning as a general base to activate the nucleophile. Another Mg2+ (MA) stabilizes the 3’-leaving group, acting as a general acid. These two metal ions exchange their roles in the second step reaction, of which MA functions as the general base and MB as the general acid.
 |
 |
References
[1] Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena.
Kruger, K., P. J. Grabowski, A. J. Zaug, J. Sands, D. E. Gottschling and T. R. Cech
Cell 31 (1): 147-57.(1982)
[2] Making ends meet: a model for RNA splicing in fungal mitochondria.
Davies, R. W., R. B. Waring, J. A. Ray, T. A. Brown and C. Scazzocchio
Nature 300 (5894): 719-24.(1982)
[3] The intervening sequence RNA of Tetrahymena is an enzyme.
Zaug, A. J. and T. R. Cech
Science 231 (4737): 470-5.(1986)
[4] Determinants of the 3’ splice site for self-splicing of the Tetrahymena pre-rRNA.
Price, J. V. and T. R. Cech
Genes Dev 2 (11): 1439-47.(1988)
[5] Compensatory mutations demonstrate that P8 and P6 are RNA secondary structure elements important for processing of a group I intron.
Williamson, C. L., N. M. Desai and J. M. Burke
Nucleic Acids Res 17 (2): 675-89.(1989)
[6] RNA structure, not sequence, determines the 5' splice-site specificity of a group I intron.
Doudna, J. A., B. P. Cormack and J. W. Szostak
Proc Natl Acad Sci U S A 86 (19): 7402-6.(1989)
[7] A conserved base pair within helix P4 of the Tetrahymena ribozyme helps to form the tertiary structure required for self-splicing.
Flor, P. J., J. B. Flanegan and T. R. Cech
EMBO J 8 (11): 3391-9.(1989)
[8] Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis.
Michel, F. and E. Westhof
J Mol Biol 216 (3): 585-610.(1990)
[9] Crystal structure of a group I ribozyme domain: principles of RNA packing.
Cate, J. H., A. R. Gooding, E. Podell, K. Zhou, B. L. Golden, C. E. Kundrot, T. R. Cech and J. A. Doudna
Science 273 (5282): 1678-85.(1996)
[10] A preorganized active site in the crystal structure of the Tetrahymena ribozyme.
Golden, B. L., A. R. Gooding, E. R. Podell and T. R. Cech
Science 282 (5387): 259-64.(1998)
[11] Crystal structure of a self-splicing group I intron with both exons.
Adams, P. L., M. R. Stahley, A. B. Kosek, J. Wang and S. A. Strobel
Nature 430 (6995): 45-50.(2004)
[12] Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site.
Guo, F., A. R. Gooding and T. R. Cech
Mol Cell 16 (3): 351-62.(2004)
[13] Crystal structure of a phage Twort group I ribozyme-product complex.
Golden, B. L., H. Kim and E. Chase
Nat Struct Mol Biol 12 (1): 82-9.(2005)
[14] Structural evidence for a two-metal-ion mechanism of group I intron splicing.
Stahley, M. R. and S. A. Strobel
Science 309 (5740): 1587-90.(2005)
[15] GISSD: Group I Intron Sequence and Structure Database.
Zhou, Y., C. Lu, Q. J. Wu, Y. Wang, Z. T. Sun, J. C. Deng and Y. Zhang
Nucleic Acids Res 36 (Database issue): D31-7.(2008)
[16] Structure-function analysis from the outside in: long-range tertiary contacts in RNA exhibit distinct catalytic roles.
Benz-Moy, T. L. and D. Herschlag
Biochemistry 50 (40): 8733-55.(2011)
[17] Cryo-EM structures of full-length Tetrahymena ribozyme at 3.1 A resolution.
Su, Z., K. Zhang, K. Kappel, S. Li, M. Z. Palo, G. D. Pintilie, R. Rangan, B. Luo, Y. Wei, R. Das and W. Chiu
Nature 596 (7873): 603-607.(2021)
[18] Sub-3-A cryo-EM structure of RNA enabled by engineered homomeric self-assembly.
Liu, D., F. A. Thelot, J. A. Piccirilli, M. Liao and P. Yin
Nat Methods 19 (5): 576-585.(2022)
 Home
Home Database
Database Research
Research About us
About us

