Group II self-splicing intron
Rfam ID: RF00029
click into different sections:
Timeline
-
1980 "Circular" introns were found to splice out from a mitochondrial gene[1]
-
1982 First secondary structure model by comparative sequence analysis[2]
-
1986 Group II introns form a lariat by self-splicing in vivo[3]
-
1986 Group II introns form a lariat by self-splicing in vivo[4]
-
1994 Common catalytic site to both splicing steps[5].
-
1995 Catalytically critical nucleotide in domain 5 [6]
-
1995 Three essential paired nucleotides in the domain 5 [7]
-
1996 Two-nucleotide bulge in D5 are important [8]
-
1997 D2 stabilizes the ribozyme core and controls the location of D6 and branching sites [10]
-
2000 Demonstration of tertiary interactions linking the catalytically critical regions of D1 to D5 and anchoring them at the 5' splice site[11]
-
2002 Crystal structures of 70-nucleotide RNAs of yeast ai5γ D5 and D6 (3 Å)[12]
-
2005 D3 is a functional group important for catalytic activity, and the interaction of D3 and D5 promotes catalysis[13]
-
2005 Single active-site region for group II intron catalysis[14]
-
2008 The first 3D structure of the Oceanobacillus iheyensis group IIC intron[16]
-
2012 Crystal structures of a group II intron at different stages of catalysis[18].
-
2014 Crystal structure of the intronic lariat form of eukaryotic group IIB[19]
-
2016 Cryo-EM structures of a group II intron in complex with its maturase[20]
-
2019 Cryo-EM structures of a group II intron reverse splicing into DNA[22]
-
2020 Two cryo-EM structures of group II intron RNPs in their pre-catalytic state[23]
Description
Group II introns are a large class of self-catalytic ribozymes and mobile genetic elements found within the genes of all three domains of life. Ribozyme activity (e.g., self-splicing) can occur under high-salt conditions in vitro. However, assistance from proteins is required for in vivo splicing. In contrast to group I introns, intron excision occurs in the absence of GTP and involves the formation of a lariat, with an A-residue branchpoint strongly resembling that found in lariats formed during splicing of nuclear pre-mRNA. It is hypothesized that pre-mRNA splicing (see spliceosome) may have evolved from group II introns, due to the similar catalytic mechanism as well as the structural similarity of the Group II Domain V substructure to the U6/U2 extended snRNA. Finally, their ability to site-specifically mobilize to new DNA sites has been exploited as a tool for biotechnology.
Structure and mechanism
2D representation
Secondary structure of the O.iheyensis intron.

|
Crystal structure of Oceanobacillus iheyensis group II intron. This representation was generated from PDB ID: 4FAR at 2.86 Å resolution.3D visualisation
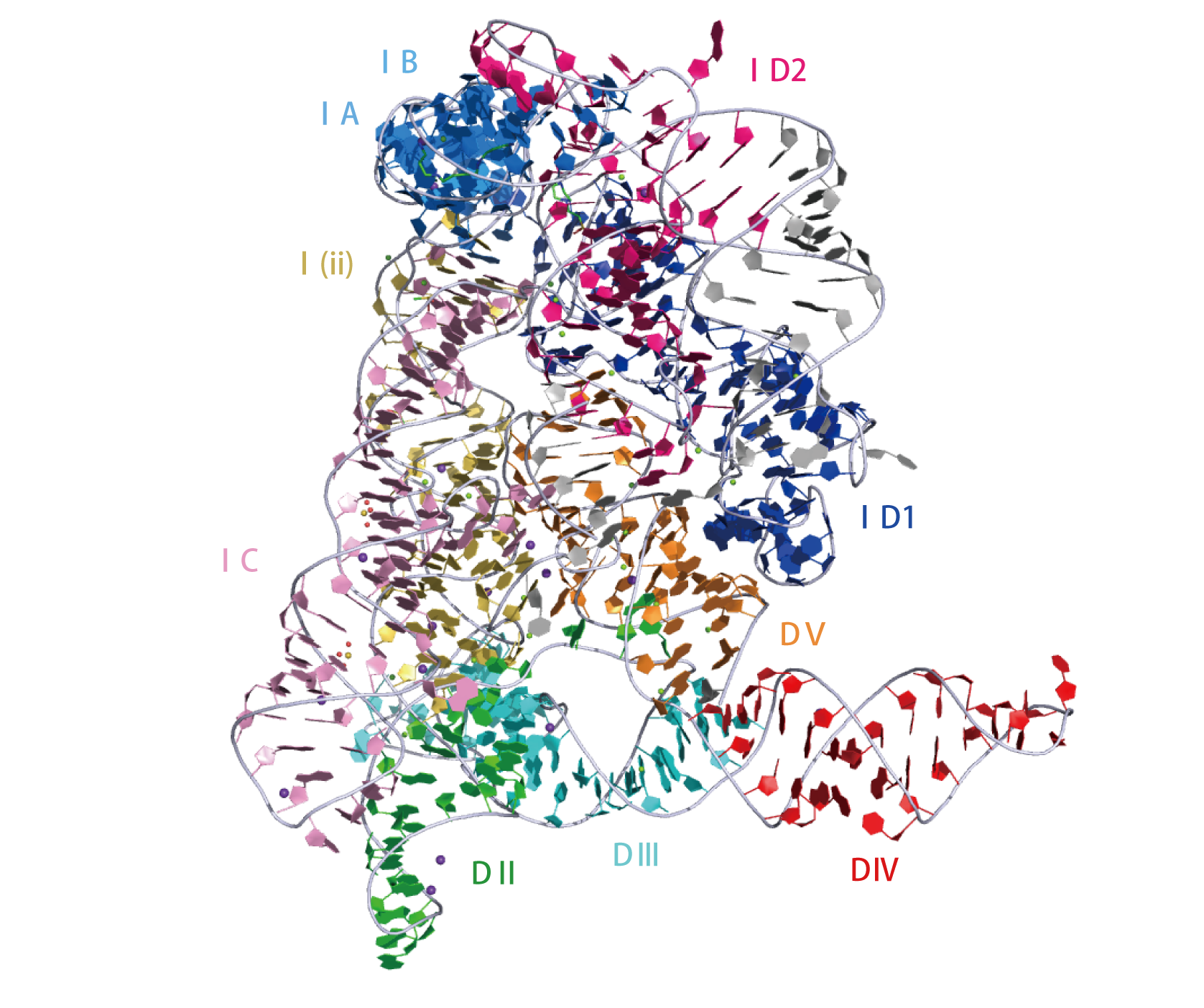 |
|
Catalytic centre
The active centre of the ribozyme. Two divalent ions (M1 and M2, green spheres) tightly coordinate the nucleotides involved in catalysis.(PDB ID: 4FAR).The transition state of first step catalysis .
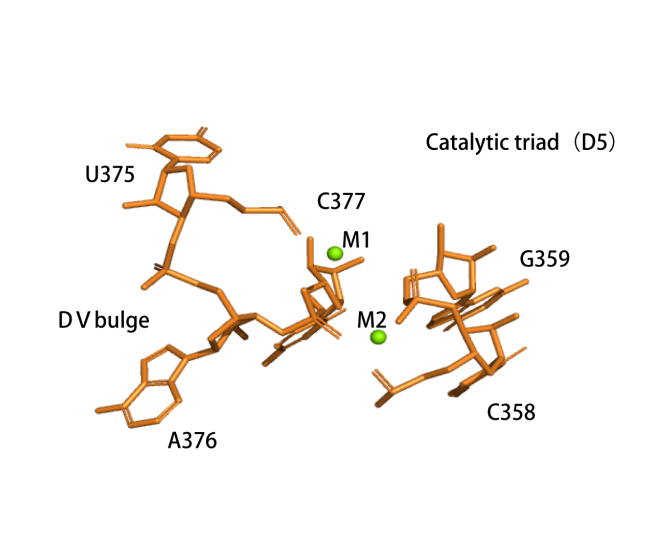 |
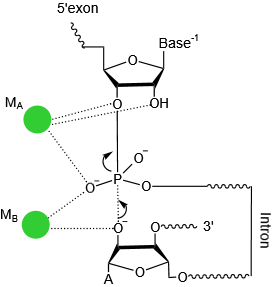 |
References
[1] A pathway of cytochrome b mRNA processing in yeast mitochondria: specific splicing steps and an intron-derived circular DNA.
Halbreich, A., P. Pajot, M. Foucher, C. Grandchamp and P. Slonimski
Cell 19 (2): 321-9.(1980)
[2] Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure.
Michel, F., A. Jacquier and B. Dujon
Biochimie 64 (10): 867-81.(1982)
[3] Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro.
van der Veen, R., A. C. Arnberg, G. van der Horst, L. Bonen, H. F. Tabak and L. A. Grivell
Cell 44 (2): 225-34.(1986)
[4] A self-splicing RNA excises an intron lariat.
Peebles, C. L., P. S. Perlman, K. L. Mecklenburg, M. L. Petrillo, J. H. Tabor, K. A. Jarrell and H. L. Cheng
Cell 44 (2): 213-23.(1986)
[5] Catalytic site components common to both splicing steps of a group II intron.
Chanfreau, G. and A. Jacquier
Science 266 (5189): 1383-7.(1994)
[6] Catalytically critical nucleotide in domain 5 of a group II intron.
Peebles, C. L., M. Zhang, P. S. Perlman and J. S. Franzen
Proc Natl Acad Sci U S A 92 (10): 4422-6.(1995)
[7] Studies of point mutants define three essential paired nucleotides in the domain 5 substructure of a group II intron.
Boulanger, S. C., S. M. Belcher, U. Schmidt, S. D. Dib-Hajj, T. Schmidt and P. S. Perlman
Mol Cell Biol 15 (8): 4479-88.(1995)
[8] Mutations of the two-nucleotide bulge of D5 of a group II intron block splicing in vitro and in vivo: phenotypes and suppressor mutations.
Schmidt, U., M. Podar, U. Stahl and P. S. Perlman
RNA 2 (11): 1161-72.(1996)
[9] Catalytic role of 2’-hydroxyl groups within a group II intron active site.
Abramovitz, D. L., R. A. Friedman and A. M. Pyle
Science 271 (5254): 1410-3.(1996)
[10] Multiple tertiary interactions involving domain II of group II self-splicing introns.
Costa, M., E. Deme, A. Jacquier and F. Michel
J Mol Biol 267 (3): 520-36.(1997)
[11] A tertiary interaction that links active-site domains to the 5’ splice site of a group II intron.
Boudvillain, M., A. de Lencastre and A. M. Pyle
Nature 406 (6793): 315-8.(2000)
[12] Structural insights into group II intron catalysis and branch-site selection.
Zhang, L. and J. A. Doudna
Science 295 (5562): 2084-8.(2002)
[13] Linking the group II intron catalytic domains: tertiary contacts and structural features of domain 3.
Fedorova, O. and A. M. Pyle
EMBO J 24 (22): 3906-16.(2005)
[14] A single active-site region for a group II intron
de Lencastre, A., S. Hamill and A. M. Pyle
Nat Struct Mol Biol 12 (7): 626-7.(2005)
[15] Group II introns: structure, folding and splicing mechanism.
Fedorova, O. and N. Zingler
Biol Chem 388 (7): 665-78.(2007)
[16] Crystal structure of a self-spliced group II intron。
Toor, N., K. S. Keating, S. D. Taylor and A. M. Pyle
Science 320 (5872): 77-82.(2008)
[17] The tertiary structure of group II introns: implications for biological function and evolution.
Pyle, A. M.
Crit Rev Biochem Mol Biol 45 (3): 215-32.(2010)
[18] Visualizing group II intron catalysis through the stages of splicing.
Marcia, M. and A. M. Pyle
Cell 151 (3): 497-507.(2012)
[19] Crystal structure of a eukaryotic group II intron lariat.
Robart, A. R., R. T. Chan, J. K. Peters, K. R. Rajashankar and N. Toor
Nature 514 (7521): 193-7.(2014)
[20] Structure of a group II intron in complex with its reverse transcriptase.
Qu, G., P. S. Kaushal, J. Wang, H. Shigematsu, C. L. Piazza, R. K. Agrawal, M. Belfort and H. W. Wang
Nat Struct Mol Biol 23 (6): 549-57.(2016)
[21] Structural Insights into the Mechanism of Group II Intron Splicing.
Zhao, C. and A. M. Pyle
Trends Biochem Sci 42 (6): 470-482.(2017)
[22] Cryo-EM Structures of a Group II Intron Reverse Splicing into DNA.
Haack, D. B., X. Yan, C. Zhang, J. Hingey, D. Lyumkis, T. S. Baker and N. Toor
Cell 178 (3): 612-623.e12.(2019)
[23] Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Liu, N., X. Dong, C. Hu, J. Zeng, J. Wang, J. Wang, H. W. Wang and M. Belfort
Nucleic Acids Res 48 (19): 11185-11198.(2020)
 Home
Home Database
Database Research
Research About us
About us

