Twister-sister
Rfam ID: RF02681
click into different sections:
Timeline
-
2015 Discovery, Secondary structure [1]
-
2017 Three-way junctional pre-catalytic structure [2]
-
2017 Four-way junctional pre-catalytic structure [3]
-
2017Structure-based mechanistic [4]
Description
The twister sister ribozyme (TS) is an RNA structure that catalyzes its own cleavage at a specific site. In other words, it is a self-cleaving ribozyme. The twister sister ribozyme was discovered by a bioinformatics strategy as an RNA Associated with Genes Associated with Twister and Hammerhead ribozymes, or RAGATH. The twister sister ribozyme has a possible structural similarity to twister ribozymes. Some striking similarities were noted, but also surprising differences, such as the absence of the two pseudoknot interactions in the twister ribozyme.
Structure and mechanism
2D representation
Secondary structure of the twister sister ribozyme with the scissile phosphates (blue) arrowed and the general acid (red) are shown. The twister sister ribozyme acts with the direct involvement of metal ions shown by green spheres with octahedral coordination of red water molecules of hydration.

|
3D visualisation
Crystal structure of the twister sister ribozyme. This representation was generated from PDB ID: 5T5A at 2.0 Å resolution. Helices are differentiated by color. T2 is a long-range tertiary interaction.
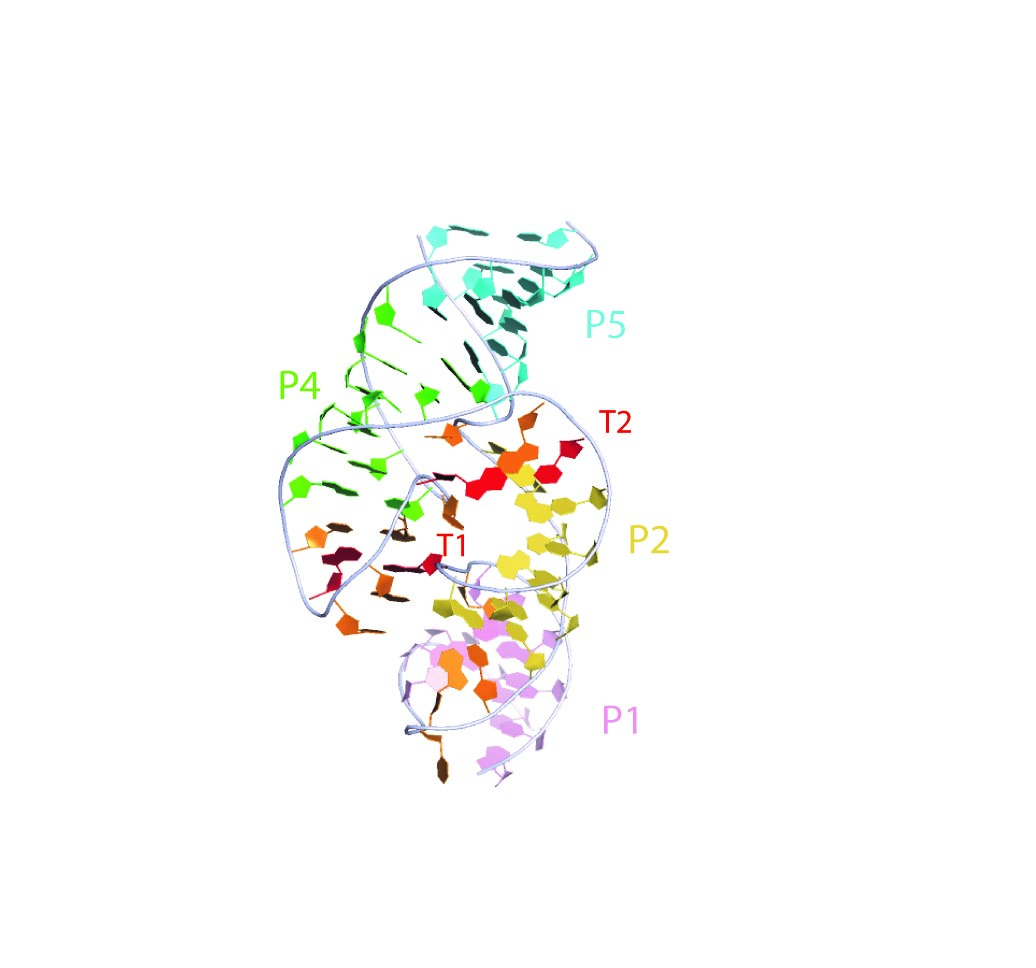 |
|
Catalytic centre
The active centre of the twister sister ribozyme. C54 and A55 flank the scissile phosphate. A metal ion (Mg) is directly bonded to C54 O2, and an inner-sphere water molecule is hydrogen bonded to A9 N3.Another inner-sphere water molecule is in position to act as the general base, shown by the connection (broken green line) to the modeled C54 O2′ nucleophile. Substitution of C7 results in a large reduction in cleavage rate, and it is likely this nucleobase is directly involved in catalysis. It is probable that this structure undergoes significant remodeling into the active geometry. Proposed mechanisms for twister sister ribozyme.Metal ions as the general base and cytosine as the general acid.
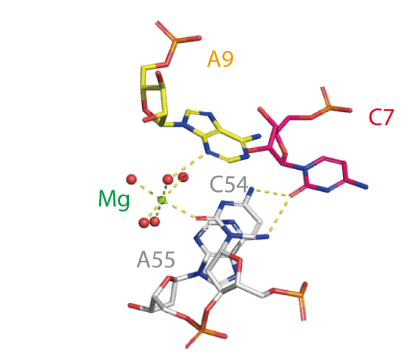 |
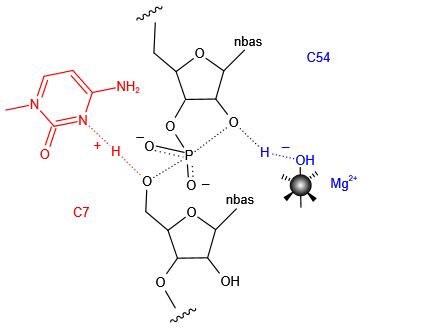 |
References
[1] New classes of self-cleaving ribozymes revealed by comparative genomics analysis.
Weinberg, Z., P. B. Kim, T. H. Chen, S. Li, K. A. Harris, C. E. Lunse and R. R. Breaker
Nat Chem Biol 11(8): 606-10.(2015)
[2] The structure of a nucleolytic ribozyme that employs a catalytic metal ion.
Liu, Y., T. J. Wilson and D. Lilley
Nat Chem Biol 13(5): 508-513.(2017)
[3] Structure-based insights into self-cleavage by a four-way junctional twister-sister ribozyme.
Zheng, L., E. Mairhofer, M. Teplova, Y. Zhang, J. Ma, D. J. Patel, R. Micura and A. Ren
Nat Commun 8(1): 1180.(2017)
[4] Structure-based mechanistic insights into catalysis by small self-cleaving ribozymes.
Ren, A., R. Micura and D. J. Patel
Curr Opin Chem Biol 41: 71-83.(2017)
[5] Model for the Functional Active State of the TS Ribozyme from Molecular Simulation.
Gaines, C. S. and D. M. York
Angew Chem Int Ed Engl 56(43): 13392-13395.(2017)
[6] Classification of the nucleolytic ribozymes based upon catalytic mechanism.
Lilley, D.
F1000Res 8.(2019)
[7] Detection of Low-Abundance Metabolites in Live Cells Using an RNA Integrator.
You, M., J. L. Litke, R. Wu and S. R. Jaffrey
Cell Chem Biol 26(4): 471-481.e3.(2019)
[8] Fundamental studies of functional nucleic acids: aptamers, riboswitches, ribozymes and DNAzymes.
Micura, R. and C. Hobartner
Chem Soc Rev 49(20): 7331-7353.(2020)
 Home
Home Database
Database Research
Research About us
About us

