Twister
click into different sections:
Timeline
-
2014 Discovery, Secondary structure[1]
-
2014 Crystal structure of P1-type[2]
-
2014 Crystal structure of P1-type [3]
-
2014 Crystal structure of P3-type [4]
-
2016 Application:Twister ribozymes as highly versatile expression platforms for artificial riboswitches[7]
-
2016 Active State of the Twister Ribozyme in Solution Predicted from Molecular Simulation[9]
-
2017 Chemical Mechanism[11]
-
2017 Mechanistic Debris Generated by Twister Ribozymes[12]
-
2021 The function of twister ribozyme variants in non-LTR retrotransposition[19]
Description
The twister ribozyme is a catalytic RNA structure capable of self-cleavage. The nucleolytic activity of this ribozyme has been demonstrated both in vivo and in vitro and has one of the fastest catalytic rates of naturally occurring ribozymes with similar function. The twister ribozyme is considered to be a member of the small self-cleaving ribozyme family which includes the hammerhead, hairpin, hepatitis delta virus (HDV), Varkud satellite (VS), and GlmS ribozymes.
Structure and mechanism
2D representation
Secondary structure of the twister ribozyme. The general acid and general base are shown in red and blue respectively.

|
3D visualisation
Crystal structure of the twister ribozyme. This representation was generated from PDB ID: 40JI at 2.34 Å resolution. Helices are differentiated by color. T1 and T2 are the two long-range tertiary interactions.
 |
|
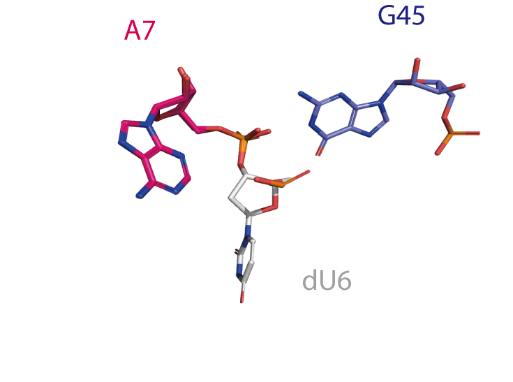
Catalytic centre
The active centre of the twister ribozyme. An O2' atom modeled onto dU6 deviates ~90° from the in-line orientation of the O2' nucleophile, P and O5' leaving group that would be optimal for activity.
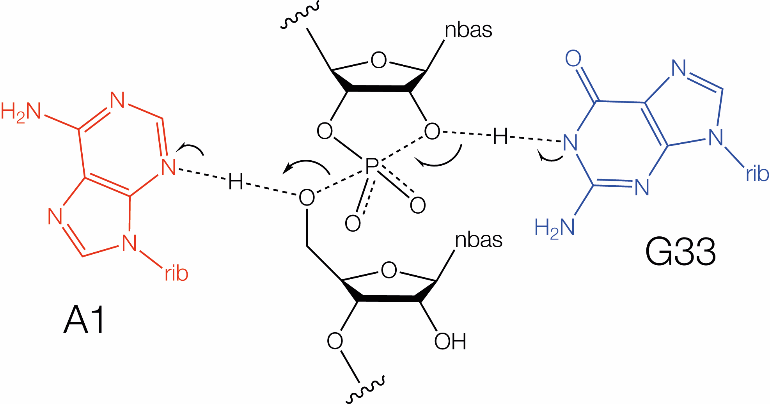
Proposed mechanisms for twister ribozyme using guanine and adenine nucleobases as general base and acid,respectively, in their
cleavage reactions.Proposed mechanisms for twister ribozyme using guanine and adenine nucleobases as general base and acid, respectively, in their cleavage reactions (A7 corresponds to A1, G33 corresponds to G45).
 |
 |
Chemical mechanism
The twister ribozyme employs general acid-base catalysis, using guanine and adenine nucleobases like the hairpin and VS ribozymes, but with some important differences. The pH dependence of cleavage rate is bell shaped, corresponding to apparent pKa values of 6.9 and 9.5 [2]. Substitutions led to the assignment of the upper pKa as resulting from G33, and like hairpin and VS, G N1 is used as the general base in cleavage. An adenine is again used as general acid in the cleavage reaction, but twister differs from the other two ribozymes in two significant respects. First, it is the highly-conserved A1, immediately 3' to the cleavage site that acts as the general acid in cleavage, and an A1G mutation led to a 104-fold loss in activity. An A1 N7C atomic mutation led to a ribozyme in which the lower pKa was shifted up by 1.5 units. In its position adjacent to the scissile phosphate it is sterically impossible to employ its N1 in proton transfer. So unusually it uses the much more acidic N3 (pKa ~ 1.5) as the general acid in the cleavage reaction [6]. However, its apparent pKa is raised by formation of two hydrogen bonds from A1 N6 to negatively-charged backbone phosphate groups. The role of A1 N3 was confirmed by atomic mutation, when it was found that the activity of A1 N3C twister ribozyme was virtually undetectable while A1 N1C twister ribozyme actually became faster.
The twister ribozyme has probably the best understood catalytic mechanism at the present time. The observed rate enhancement arises from four contributions [6] :
1. The structure observed in the crystal may be readily remodeled into a conformation that is well aligned for SN2 attack of the O2' nucleophile on the scissile phosphate in the cleavage reaction.
2. The transition state is stabilized by accepting a hydrogen bond from G33 N2.
3. G33 N1 removes a proton from the O2' to make it a stronger nucleophile.
4. A1 N3 donates a proton to the O5' oxyanion leaving group.
References
[1] A widespread self-cleaving ribozyme class is revealed by bioinformatics.
Roth, A., Z. Weinberg, A. G. Chen, P. B. Kim, T. D. Ames and R. R. Breaker
Nat Chem Biol 10(1): 56-60.(2014)
[2] Crystal structure and mechanistic investigation of the twister ribozyme.
Liu, Y., T. J. Wilson, S. A. McPhee and D. M. Lilley
Nat Chem Biol 10(9): 739-44.(2014)
[3] In-line alignment and Mg(2)(+) coordination at the cleavage site of the env22 twister ribozyme.
Ren, A., M. Kosutic, K. R. Rajashankar, M. Frener, T. Santner, E. Westhof, R. Micura and D. J. Patel
Nat Commun 5: 5534.(2014)
[4] Structural basis for the fast self-cleavage reaction catalyzed by the twister ribozyme.
Eiler, D., J. Wang and T. A. Steitz
Proc Natl Acad Sci U S A 111(36): 13028-33.(2014)
[5] A Mini-Twister Variant and Impact of Residues/Cations on the Phosphodiester Cleavage of this Ribozyme Class.
Kosutic, M., S. Neuner, A. Ren, S. Flur, C. Wunderlich, E. Mairhofer, N. Vusurovic, J. Seikowski, K. Breuker, C. Hobartner, D. J. Patel, C. Kreutz and R. Micura
Angew Chem Int Ed Engl 54(50): 15128-15133.(2015)
[6] The Novel Chemical Mechanism of the Twister Ribozyme.
Wilson, T. J., Y. Liu, C. Domnick, S. Kath-Schorr and D. M. Lilley
J Am Chem Soc 138(19): 6151-62.(2016)
[7] Twister ribozymes as highly versatile expression platforms for artificial riboswitches.
Felletti, M., J. Stifel, L. A. Wurmthaler, S. Geiger and J. S. Hartig
Nat Commun 7: 12834.(2016)
[8] High-Throughput Mutational Analysis of a Twister Ribozyme.
Kobori, S. and Y. Yokobayashi
Angew Chem Int Ed Engl 55(35): 10354-7.(2016)
[9] Ribozyme Catalysis with a Twist: Active State of the Twister Ribozyme in Solution Predicted from Molecular Simulation.
Gaines, C. S. and D. M. York
J Am Chem Soc 138(9): 3058-65.(2016)
[10] Pseudoknot Formation Seeds the Twister Ribozyme Cleavage Reaction Coordinate.
Vusurovic, N., Altman, R. B., Terry, D. S., Micura, R. and Blanchard, S. C.
J Am Chem Soc 139 (24): 8186-8193.(2017)
[11] Unwinding the twister ribozyme: from structure to mechanism.
Gebetsberger, J. and Micura, R.
Wiley Interdiscip Rev RNA 8 (3).(2017)
[12] Mechanistic Debris Generated by Twister Ribozymes.
Breaker, R. R.
ACS Chem Biol 12(4): 886-891.(2017)
[13] Metals induce transient folding and activation of the twister ribozyme.
Panja, S., B. Hua, D. Zegarra, T. Ha and S. A. Woodson
Nat Chem Biol 13(10): 1109-1114.(2017)
[14] Cellular Small Molecules Contribute to Twister Ribozyme Catalysis.
Messina, K. J. and P. C. Bevilacqua
J Am Chem Soc 140(33): 10578-10582.(2018)
[15] Cleaning Up Mechanistic Debris Generated by Twister Ribozymes Using Computational RNA Enzymology.
Gaines, C. S., T. J. Giese and D. M. York
ACS Catal 9(7): 5803-5815.(2019)
[16] Classification of the nucleolytic ribozymes based upon catalytic mechanism.
Lilley, D.
F1000Res 8.(2019)
[17] Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts.
Litke, J. L. and S. R. Jaffrey
Nat Biotechnol 37(6): 667-675.(2019)
[18] Light-controlled twister ribozyme with single-molecule detection resolves RNA function in time and space.
Korman, A., H. Sun, B. Hua, H. Yang, J. N. Capilato, R. Paul, S. Panja, T. Ha, M. M. Greenberg and S. A. Woodson
Proc Natl Acad Sci U S A 117(22): 12080-12086.(2020)
[19] The function of twister ribozyme variants in non-LTR retrotransposition in Schistosoma mansoni.
Liu, G., H. Jiang, W. Sun, J. Zhang, D. Chen and A. Murchie
Nucleic Acids Res 49(18): 10573-10588.(2021)
 Home
Home Database
Database Research
Research About us
About us

